User:Estelle Blochouse/ Sandbox 1497
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Function == | == Function == | ||
| - | Multicopper oxidases are enzymes involved in copper homeostasis. | + | Multicopper oxidases are enzymes involved in copper homeostasis. Indeed, free copper present in a cell (so, not bounded to a protein) is harmful and can cause cellular damage. It needs to be regulated. |
Multicopper oxidase acts probably for the detoxification of copper present in the periplasmic space. It oxidizes the Cu+ into Cu2+ and prevents its uptake by the cytoplasm. It also possesses a phenoloxidase and a ferroxidase activity which can be involved in the prevention of oxidative damage.<ref><span class='plainlinks'>[https://www.uniprot.org/uniprot/P36649 UniProtKB]</span></ref> | Multicopper oxidase acts probably for the detoxification of copper present in the periplasmic space. It oxidizes the Cu+ into Cu2+ and prevents its uptake by the cytoplasm. It also possesses a phenoloxidase and a ferroxidase activity which can be involved in the prevention of oxidative damage.<ref><span class='plainlinks'>[https://www.uniprot.org/uniprot/P36649 UniProtKB]</span></ref> | ||
| Line 13: | Line 13: | ||
Multicopper oxidase might also be involved in the regulation of metal transport. | Multicopper oxidase might also be involved in the regulation of metal transport. | ||
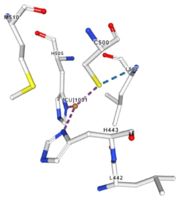
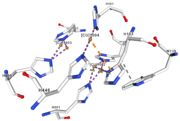
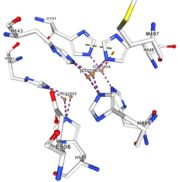
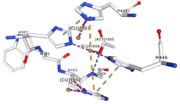
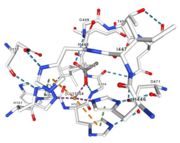
| - | Multicopper oxidases are able to oxidise their substrates thanks to their particular structure. | + | Multicopper oxidases are able to oxidise their substrates thanks to their particular structure. They own four copper ions (Cu1001, Cu1002, Cu1003 and C1004) dispatched between two important areas : the mononuclear copper center (Cu1001) and the trinuclear copper center (Cu1002, Cu1003 and Cu1004). |
| - | They accept an electron in the <scene name='pdbligand=CU:COPPER+(II)+ION'>mononoclear copper center</scene> and transfer it to the trinuclear copper | + | They accept an electron in the <scene name='pdbligand=CU:COPPER+(II)+ION'>mononoclear copper center</scene> and transfer it to the trinuclear copper center. The dioxygen binds to the trinuclear center and receives four electrons. It is transformed into two molecules of water.<ref>Bento I, Martins LO, Gato Lopes G, Arménia Carrondo M, Lindley PF, Dioxygen reduction by multi-copper oxidases; a structural perspective, November 2005</ref> Three differents copper centres exist that can be differentiated spectroscopically: Type 1 or blue (<scene name='pdbligand=CU:COPPER+(II)+ION'>mononoclear copper center</scene>)(Cu1001), type 2 or normal (Cu1004) and type 3 or coupled binuclear (Cu1002 and Cu1003).<ref>Messerschmidt A, Huber R, The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships, Eur. J. Biochem. 187, January 1990</ref><ref>Ouzounis C, Sander C, A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins, FEBS Lett. volume 279, February 1991</ref> |
== Mechanism == | == Mechanism == | ||
Revision as of 18:10, 11 January 2019
Multicopper Oxidase CueO (4e9s)
| |||||||||||
References
- ↑ EMBL-EBI, Family: Cu-oxidase (PF00394), Summary: Multicopper oxidase, http://pfam.xfam.org/family/Cu-oxidase
- ↑ UniProtKB
- ↑ Bento I, Martins LO, Gato Lopes G, Arménia Carrondo M, Lindley PF, Dioxygen reduction by multi-copper oxidases; a structural perspective, November 2005
- ↑ Messerschmidt A, Huber R, The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships, Eur. J. Biochem. 187, January 1990
- ↑ Ouzounis C, Sander C, A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins, FEBS Lett. volume 279, February 1991
- ↑ Hirofumi Komori, Ryosuke Sugiyama, Kunishige Kataoka, Kentaro Miyazaki, Yoshiki Higuchib, and Takeshi Sakurai, New insights into the catalytic active-site structure of multicopper oxidases, Biological Crystallography, 6 December 2013 doi:10.1107/S1399004713033051
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ RCBS PDB
- ↑ Kataoka K, Komori H, Ueki Y, Konno Y, Kamitaka Y, Kurose S, Tsujimura S, Higuchi Y, Kano K, Seo D, Sakurai T. Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site. J Mol Biol. 2007 Oct 12;373(1):141-52. Epub 2007 Aug 2. PMID:17804014 doi:10.1016/j.jmb.2007.07.041