Sandbox Reserved 1500
From Proteopedia
| Line 4: | Line 4: | ||
<table><tr><td colspan='2'>[[5hfg]] is a 1 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5HFG OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5HFG FirstGlance]. <br> | <table><tr><td colspan='2'>[[5hfg]] is a 1 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5HFG OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5HFG FirstGlance]. <br> | ||
| - | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[5hfi|5hfi]]</td></tr> | |
<tr id='related'><td class="sblockLbl"><b>Descriptor:</b></td><td class="sblockDat">Uncharacterized protein, cytosolic disulfide reductase DsbM</td></tr> | <tr id='related'><td class="sblockLbl"><b>Descriptor:</b></td><td class="sblockDat">Uncharacterized protein, cytosolic disulfide reductase DsbM</td></tr> | ||
| - | + | <tr id='related'><td class="sblockLbl"><b>Functional Keywords:</b></td><td class="sblockDat">5hfi|dsb, dsbm, disulfide reductase, thioredoxin superfamilly, oridoreductase</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Biological source:</b></td><td class="sblockDat">Pseudomonas aeruginosa PAO1</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Total number of polymer chains:</b></td><td class="sblockDat">1</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Total molecular weight:</b></td><td class="sblockDat">25,286.7 Da</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Authors:</b></td><td class="sblockDat">Jo. I., Ha, N.-C. (release date : 2016-10-26)</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Experimental method:</b></td><td class="sblockDat">X-Ray diffraction (1.823Å) Those informations are from [http://www.example.com PDBJ].</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Global symmetry:</b></td><td class="sblockDat">Asymmetric - C1</td></tr> | |
| - | + | <tr id='related'><td class="sblockLbl"><b>Global stoichiometry:</b></td><td class="sblockDat">Monomer - A</td></tr> | |
<tr id='ligand'><td class="sblockLbl"><b>Secondary structure:</b></td><td class="sblockLbl">Beta sheets[[Image:5hfgplan.PNG|thumb|middle|'''Figure 1:''' 5HFG secondary structure.<ref>PDB image</ref>]], alpha helix</td></tr> | <tr id='ligand'><td class="sblockLbl"><b>Secondary structure:</b></td><td class="sblockLbl">Beta sheets[[Image:5hfgplan.PNG|thumb|middle|'''Figure 1:''' 5HFG secondary structure.<ref>PDB image</ref>]], alpha helix</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5hfg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5hfg OCA], [http://pdbe.org/5hfg PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=5hfg RCSB], [http://www.ebi.ac.uk/pdbsum/5hfg PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=5hfg ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5hfg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5hfg OCA], [http://pdbe.org/5hfg PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=5hfg RCSB], [http://www.ebi.ac.uk/pdbsum/5hfg PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=5hfg ProSAT]</span></td></tr> | ||
Revision as of 18:35, 11 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
|
Contents |
Structural highlights
| 5hfg is a 1 chain structure. Full crystallographic information is available from OCA. For a guided tour on the structure components use FirstGlance. | |
| Related: | 5hfi |
| Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM |
| Functional Keywords: | 5hfi|dsb, dsbm, disulfide reductase, thioredoxin superfamilly, oridoreductase |
| Biological source: | Pseudomonas aeruginosa PAO1 |
| Total number of polymer chains: | 1 |
| Total molecular weight: | 25,286.7 Da |
| Authors: | Jo. I., Ha, N.-C. (release date : 2016-10-26) |
| Experimental method: | X-Ray diffraction (1.823Å) Those informations are from PDBJ. |
| Global symmetry: | Asymmetric - C1 |
| Global stoichiometry: | Monomer - A |
| Secondary structure: | Beta sheets Figure 1: 5HFG secondary structure.[1] |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum, ProSAT |
Primary structure
5hfg is a 1 chain structure of 238 amino acids. [2]
Secondary structure
The structure of 5hfg mainly consists in alpha helix (12) , beta sheets (4), you can check the 3D view here. 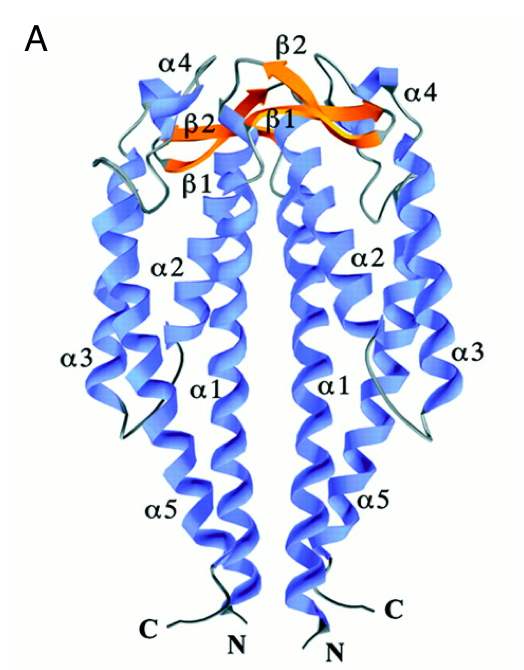
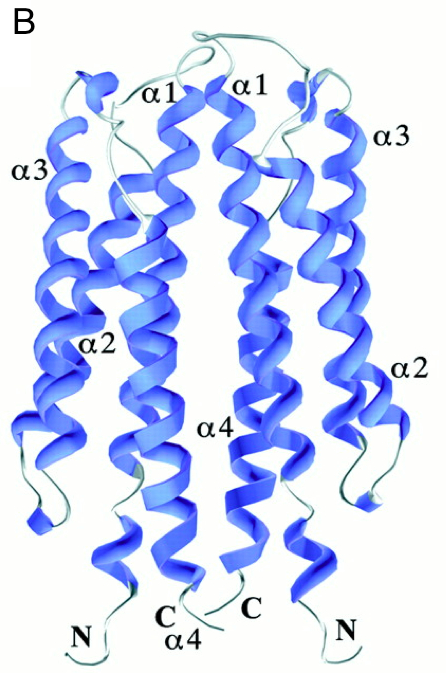
It has 2 main domains, the lid domain and the thioredoxine domain (see the picture). 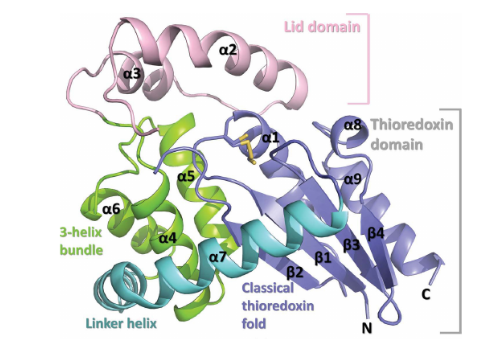
Tertiary structure
5hfg behaves as a monomer or a dimer in solution but it has a monomeric composition. 5hfg is one distinct polypeptide molecule. [3]
Nature
5hfg is a disulfide reductase DsbM from Pseudomonas aeruginosa. The Dsb family of proteins is mainly used to oxidize and reduce cysteine residues of substrate proteins. Most enzymes from Dsb-family catalyze disulfide formation in periplasmic or secreted substrate proteins. [4]
It has no bound ligands and no modified residues. Sequence domains: Thioredoxin-like superfamily DSBA-like thioredoxin domain
Function
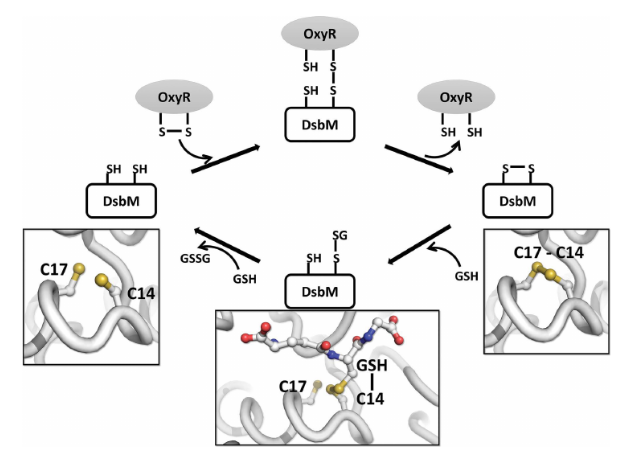
This protein has a disulfide oxidoreductase activity, it is implicated in the reduction of the cytoplasmicredox-sensor protein OxyR in Pseudomonas aeruginosa. It contains a CXXC motif which sequence is homologous to the Dsb-family proteins. The CXXC motif is the actif site of the protein for the catalysis of disulfide oxidoreduction, moreover, this site also caracterize the affinity with the substrate protein. 5hfg presents a CGWC sequence in its CXXC motif, in which the Trp16 residue facilitates some hydrophobic interactions between the lid domain and the thioredoxine domain.[5]
According to biochemical studies, 5hfg have also an ability to reduce the disulfide of OxyR (Hydrogen peroxide-inducible genes activator) that is formed by H2O2. The following photo shows a proposed mechanism of the action of DsbM in which the oxidized OxyR and the reduced glutathione are transported to the glutathione binding site of DsbM.
References
- ↑ PDB image
- ↑ https://www.rcsb.org/structure/5HFG
- ↑ https://www.ebi.ac.uk/pdbe/entry/pdb/5HFG
- ↑ https://www.researchgate.net/publication/308186632_Crystal_structures_of_the_disulfide_reductase_DsbM_from_Pseudomonas_aeruginosa
- ↑ https://www.researchgate.net/publication/308186632_Crystal_structures_of_the_disulfide_reductase_DsbM_from_Pseudomonas_aeruginosa