We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Frataxin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
<StructureSection load='2fql' size='340' side='right' caption='Yeast frataxin (PDB code [[2fql]])' scene='78/788815/Spacefill_model/1'> | <StructureSection load='2fql' size='340' side='right' caption='Yeast frataxin (PDB code [[2fql]])' scene='78/788815/Spacefill_model/1'> | ||
| Line 26: | Line 25: | ||
The sequences comprising each beta-sheet (in <font color='darkorchid'><b>purple</b></font>) are the following: Pro 100-Ser 105 for <scene name='78/788815/Beta_1/1'>beta 1</scene>, Val 108-Ile 113 for <scene name='78/788815/Beta_2/1'>beta 2</scene>, Gln 124-Gly 117 for <scene name='78/788815/Beta_3/1'>beta 3</scene>, Ser 134-Gln 129 for <scene name='78/788815/Beta_4/1'>beta 4</scene>, Asp 143-Gly 138 for <scene name='78/788815/Beta_5/1'>beta 5</scene>, Trp 149-Leu 152 for <scene name='78/788815/Beta_6/1'>beta 6</scene> and the short region between Lys 157-Gly 155 for <scene name='78/788815/Beta_7/1'>beta 7</scene>. | The sequences comprising each beta-sheet (in <font color='darkorchid'><b>purple</b></font>) are the following: Pro 100-Ser 105 for <scene name='78/788815/Beta_1/1'>beta 1</scene>, Val 108-Ile 113 for <scene name='78/788815/Beta_2/1'>beta 2</scene>, Gln 124-Gly 117 for <scene name='78/788815/Beta_3/1'>beta 3</scene>, Ser 134-Gln 129 for <scene name='78/788815/Beta_4/1'>beta 4</scene>, Asp 143-Gly 138 for <scene name='78/788815/Beta_5/1'>beta 5</scene>, Trp 149-Leu 152 for <scene name='78/788815/Beta_6/1'>beta 6</scene> and the short region between Lys 157-Gly 155 for <scene name='78/788815/Beta_7/1'>beta 7</scene>. | ||
| - | Concerning the '''tertiary structure''', each subunit has a folding pattern called α/β sandwich, in | + | Concerning the '''tertiary structure''', each subunit has a folding pattern called α/β sandwich, in which two alpha-helices are packed against five strands of anti-parallel β-sheets. <scene name='78/788815/Ab_folding/1'>Click here</scene> to see it in the whole trimeric form. |
---- | ---- | ||
---- | ---- | ||
| - | + | ==Stabilization of the Trimeric Quaternary Structure== | |
| - | The trimeric structure of frataxin consists of the association of three monomers, and is mainly stabilized by the <scene name='78/788815/Stabilization_of_trimer/1'>N-terminal extensions</scene> of each subunit, shown in <span style="color:yellow;background-color:darkgrey;font-weight:bold;">yellow</span>. These consist of loops, with a short helical N-terminal segment (alpha-helix 1; recall its secondary structure) highly flexible in the monomer solution, but interestingly, when in the trimeric | + | The trimeric structure of frataxin consists of the association of three monomers, and is mainly stabilized by the <scene name='78/788815/Stabilization_of_trimer/1'>N-terminal extensions</scene> of each subunit, shown in <span style="color:yellow;background-color:darkgrey;font-weight:bold;">yellow</span>. These consist of loops, with a short helical N-terminal segment (alpha-helix 1; recall its secondary structure) highly flexible in the monomer solution, but interestingly, when in the trimeric arrangement, they play a crucial role in maintaining it. Viewing <scene name='78/788815/Stabilization_of_trimer_back/2'>the other side</scene> of the molecule, we can notice how the N-terminal extensions, still in <span style="color:yellow;background-color:darkgrey;font-weight:bold;">yellow</span>, interact with the <font color='rosybrown'><b>adjacent monomer</b></font>. Taking a <scene name='78/788815/Stabilization_of_trimer_zoom_1/3'>closer look</scene>, it is possible figure out how the N-terminal loop of the first monomer, here described as chain A, is placed with respect to chain B. |
<scene name='78/788815/Stabilization_of_trimer_resid1/4'>Exploring even further</scene> the details, it is possible to see some residues close enough to interact. The names associated with their positions can be seen by <scene name='78/788815/All_residues_at_end/1'>clicking here</scene>. | <scene name='78/788815/Stabilization_of_trimer_resid1/4'>Exploring even further</scene> the details, it is possible to see some residues close enough to interact. The names associated with their positions can be seen by <scene name='78/788815/All_residues_at_end/1'>clicking here</scene>. | ||
| - | <scene name='78/788815/All_residues_at_end_transparen/3'>Click here</scene> to rotate this region. We can <scene name='78/788815/All_residues_at_end_transp_hyd/1'>color</scene> the residues differently according to their hydrophilicity. In this new color scheme, <font color='fuchsia'><b>polar residues are represented in pink</b></font> while the <font color='darkgrey'><b>hydrophobic ones appear gray</b></font>. Now we are about to <scene name='78/788815/All_residues_at_end_transp_tur/1'>color</scene> all those relevant residues to specify their interactions. The <scene name='78/788815/All_residues_at_end_transp_pac/1'>package of hydrophobic residues</scene> can be seen. Here,<font color='navy'><b> Pro 62, Val 65 and Leu 68, shown in dark-blue</b></font>, are packed against the <font color='red'><b>polar uncharged aminoacids Thr 110 and Thr 118, in red</b></font> (other | + | <scene name='78/788815/All_residues_at_end_transparen/3'>Click here</scene> to rotate this region. We can <scene name='78/788815/All_residues_at_end_transp_hyd/1'>color</scene> the residues differently according to their hydrophilicity. In this new color scheme, <font color='fuchsia'><b>polar residues are represented in pink</b></font> while the <font color='darkgrey'><b>hydrophobic ones appear gray</b></font>. Now we are about to <scene name='78/788815/All_residues_at_end_transp_tur/1'>color</scene> all those relevant residues to specify their interactions. The <scene name='78/788815/All_residues_at_end_transp_pac/1'>package of hydrophobic residues</scene> can be seen. Here,<font color='navy'><b> Pro 62, Val 65 and Leu 68, shown in dark-blue</b></font>, are packed against the <font color='red'><b>polar uncharged aminoacids Thr 110 and Thr 118, in red</b></font> (other amino acids are shown in <font color='mediumturquoise'><b>turquoise</b></font>). This interaction among the hydrophobic residues contributes to the maneintance of the loop configuration of the N-terminal region at its extremity. Another important interaction is the <scene name='78/788815/All_residues_at_end_transp_bon/1'>hydrogen bond</scene> formed between <font color='orangered'><b>Glu 64</b></font> and <font color='blueviolet'><b>Thr 118</b></font>. Those are the only residues able to form hydrogen bond, since the <scene name='78/788815/Hydrogen_bond_n-term-correct/3'>distance separating them</scene> is within a range of approximately 3 Å (or 0.3 nm). <scene name='78/788815/Hydrogen_bond_n-term-correct/4'>In this image</scene>, pay special attention it the role of the <font color='red'><b>carbonyl oxygen</b></font> of Glu 64 involved in the hydrogen bonding. In this color scheme, <font color='grey'><b> carbons are grey</b></font>, <font color='red'><b>oxygens are red</b></font> and <font color='blue'><b>nitrogens are blue</b></font>. |
Now, we can devote our attention to examine what occurs at the <scene name='78/788815/Stabilization_of_trimer_base/1'>base of the N-terminal region</scene>.Those are the <scene name='78/788815/Residues_at_base_-_2/1'>residues involved</scene> in relevant interactions that contribute to the stabilization of the trimeric form. Those are their specific <scene name='78/788815/Residues_at_base_-_names/1'>names</scene> (different colors of the labels simply indicates different subunits). <scene name='78/788815/Names-transparent/1'>Click here</scene> to give emphasis on them, and <scene name='78/788815/Names-transparent-zoom-clear/1'>here</scene> to get a better spatial notion of its arrangement. | Now, we can devote our attention to examine what occurs at the <scene name='78/788815/Stabilization_of_trimer_base/1'>base of the N-terminal region</scene>.Those are the <scene name='78/788815/Residues_at_base_-_2/1'>residues involved</scene> in relevant interactions that contribute to the stabilization of the trimeric form. Those are their specific <scene name='78/788815/Residues_at_base_-_names/1'>names</scene> (different colors of the labels simply indicates different subunits). <scene name='78/788815/Names-transparent/1'>Click here</scene> to give emphasis on them, and <scene name='78/788815/Names-transparent-zoom-clear/1'>here</scene> to get a better spatial notion of its arrangement. | ||
| - | If we <scene name='78/788815/Residues_at_base_-_2_polarity/1'>color according to their polarities</scene> (recall: <font color='fuchsia'><b>pink</b></font> for charged aminoacids, and <font color='darkgrey'><b>grey</b></font> for aliphatic ones), it becomes evident their charged nature. Them, there is no hydrophobic packing taking place at this region. Instead, there are '''hydrogen bonds''' as the main eletrostatic interaction. Notice, again, the <scene name='78/788815/Names-transparent-elements/2'>element composition</scene> of each | + | If we <scene name='78/788815/Residues_at_base_-_2_polarity/1'>color according to their polarities</scene> (recall: <font color='fuchsia'><b>pink</b></font> for charged aminoacids, and <font color='darkgrey'><b>grey</b></font> for aliphatic ones), it becomes evident their charged nature. Them, there is no hydrophobic packing taking place at this region. Instead, there are '''hydrogen bonds''' as the main eletrostatic interaction. Notice, again, the <scene name='78/788815/Names-transparent-elements/2'>element composition</scene> of each amino acid: in this color scheme, again, we have {{Template:ColorKey_Element_C}}, {{Template:ColorKey_Element_O}}, and {{Template:ColorKey_Element_N}}. |
There are six hydrogen-bonding pairs contributing to the stabilization of the molecule. Between <scene name='78/788815/His_74_lys_72/1'>Lys 72 and His 74</scene>, <scene name='78/788815/His_74_and_glu_76/1'>His 74 and Glu 76</scene> and <scene name='78/788815/Asp_78_and_glu_75/1'>Glu 75 and Asp 78</scene>, the pair is always formed between the <font color='red'><b>carbonyls of the first</b></font> and the <font color='blue'><b>amide groups of the second</b></font>. | There are six hydrogen-bonding pairs contributing to the stabilization of the molecule. Between <scene name='78/788815/His_74_lys_72/1'>Lys 72 and His 74</scene>, <scene name='78/788815/His_74_and_glu_76/1'>His 74 and Glu 76</scene> and <scene name='78/788815/Asp_78_and_glu_75/1'>Glu 75 and Asp 78</scene>, the pair is always formed between the <font color='red'><b>carbonyls of the first</b></font> and the <font color='blue'><b>amide groups of the second</b></font>. | ||
| - | The other type of hydrogen bonding occurs between the side chains of the | + | The other type of hydrogen bonding occurs between the side chains of the amino acids involved in the pairs <scene name='78/788815/His_74_asp_79/1'>His 74 and Asp 79</scene>, <scene name='78/788815/Asp_78_and_lys_123/1'>Asp 78 and Lys 123</scene> and <scene name='78/788815/Glu76-arg141/2'>Glu76 and Arg141</scene>. In any of each ways, there is always an <font color='red'><b>oxygen</b></font> and a <font color='blue'><b>nitrogen</b></font> involved. |
---- | ---- | ||
| Line 49: | Line 48: | ||
| - | + | ==The Role of the Channel== | |
The channel of the trimer is formed at its 3-fold axis, in a central position. It is the strure responsible for stabilizing the atom of iron for delivery, but also acts to further stabilise the trimeric form. | The channel of the trimer is formed at its 3-fold axis, in a central position. It is the strure responsible for stabilizing the atom of iron for delivery, but also acts to further stabilise the trimeric form. | ||
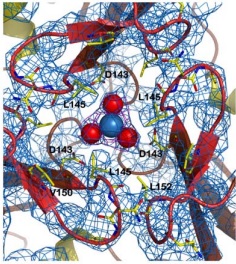
| - | + | Stabilizing one side of the channel, there are some interactions between the <scene name='78/786054/151-155_144-149/1'>loops around the central channel</scene>, which form a helical structure maintained, for instance, through hydrogen bonds between the pairs <scene name='78/786054/152-147_hydrogen_bond/1'>Leu 152-Gly 147</scene> and <scene name='78/786054/146-154_hydrogen_bond/1'>Asn 146-Gln 154</scene>. this last one also stabilizes the ASN146 interaction with the iron atom, as described in the figure below | |
| - | In the structure of the channel, the side chains of the hydrophobic | + | In the structure of the channel, the side chains of the hydrophobic amino acids Leu 145, Val 150 and Leu 152 are exposed to the solvent, creating a <scene name='78/786054/Hydrophobic_lid/3'>hydrophobic lid</scene> around the channel. This hydrophobic lid isolates one of the sides of the channel core, as well as providing a hydrophobic contact surface by which other proteins can interact, hiding their hydrophobic residues from the solvent-rich environment. |
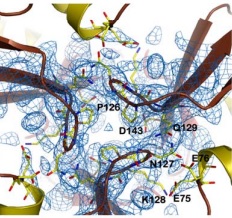
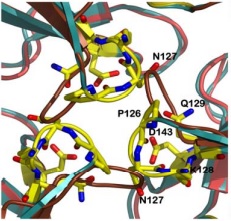
| - | Other structure in the central channel of high importance is the loop formed by <scene name='78/786054/125-128/3'>125-128</scene>. This loop, | + | Other structure in the central channel of high importance is the loop formed by <scene name='78/786054/125-128/3'>125-128</scene>. This loop, stabilized by the hydrogen bonds between <scene name='78/786054/143-129__hydrogen_bond/1'>Asp 143-Gln 129</scene>, <scene name='78/786054/Bound_127-75/1'>127-75</scene> and <scene name='79/790348/129-126/1'>129-126</scene>, is present in 2 different conformations. The one just described is involved in further stabilizing the Fe atom within the channel. The metal ion binds at around 4 Å from the side chains of the <scene name='78/788815/Iron_channel/1'>three Asp 143 residues</scene> (distances between residues are shown for reference). Laboratory data from X-ray crystallography suggests the Fe 2+ ion being associated with solvent molecules. In this conformation the PRO126 in pointing toward the center of the channel, further reducing contact of the iron with the environment. This create an <scene name='78/786054/Fe_isolation_chamber/1'>isolation chamber</scene> for the iron inside the channel, increasing the stability for the transportation of iron. |
[[Image:ironload.jpg]][[Image:conf1.jpg]] | [[Image:ironload.jpg]][[Image:conf1.jpg]] | ||
| - | the other conformation that the 125-128 loop can take is were it fold outside of the channel, | + | the other conformation that the 125-128 loop can take is were it fold outside of the channel, exhibiting interactions with the residues N127-Q129-D143. in this conformations, the hydrogens interactions between N127-E75 and Q129-P126 are not present. |
[[Image:conf2.jpg]] | [[Image:conf2.jpg]] | ||
| Line 75: | Line 74: | ||
== Disease == | == Disease == | ||
| - | Friedreich ataxia is developed when the frataxin trimer cannot | + | Friedreich ataxia is developed when the frataxin trimer cannot stabilize itself to perform the ferroxidase activity, with is probably dependent on conformational changes that take place when the monomers come together, being in a site around the residues H74, D78, D79, D82 and H83. |
| - | There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, | + | There are 4 know mutations capable of creating such defects: <scene name='78/786054/Mutation_107_123/1'>G107V</scene>, that breaks the hydrogen bond between G107 and K123, destabilizing the region of 123-130; W131, necessary to stabilize the N-terminal extension; <scene name='78/786054/Mutation_141-125/1'>R141C</scene> breaks an hydrogen bound between R141-P125, which may affect the conformation of the 125-128 loop and D122Y |
</StructureSection> | </StructureSection> | ||
| + | == 3D Structures of frataxin == | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Frataxin | ||
| + | |||
| + | **[[1ekg]], [[3s4m]] – hFXN C-terminal - human<br /> | ||
| + | **[[1ly7]] – hFXN C-terminal - NMR<br /> | ||
| + | **[[3s5d]], [[3s5e]], [[3s5f]], [[3t3j]], [[3t3k]], [[3t3l]], [[3t3t]], [[3t3x]] – hFXN C-terminal (mutant)<br /> | ||
| + | **[[5kz5]] – hFXN C-terminal + iron sulfur assembly protein + cysteine desulfarase – Cryo EM<br /> | ||
| + | |||
| + | *FXN homolog; yeast residues 53-174 | ||
| + | |||
| + | **3co6, 3coa, [[3co7]], [[2ga5]] – yFH1 – yeast – NMR<br /> | ||
| + | **[[2fql]], [[3oeq]] – yFH1 (mutant)<br /> | ||
| + | **[[3oer]] – yFH1 (mutant) + Co<br /> | ||
| + | **[[4ec2]] – yFH1 (mutant) + Fe<br /> | ||
| + | **[[5tre]], [[5t0v]] – yFH1 + iron sulfur assembly protein – Cryo EM<br /> | ||
| + | |||
| + | *FXN ortholog CyaA | ||
| + | |||
| + | **[[1ew4]] – EcCyaY – Escherichia coli <br /> | ||
| + | **[[1soy]] – EcCyaY – NMR<br /> | ||
| + | **[[2eff]] – EcCyaY + Co <br /> | ||
| + | **[[2p1x]] – EcCyaY + Eu <br /> | ||
| + | **[[4jpd]] – CyaY – Burkholderia cenocepacia<br /> | ||
| + | **[[4hs5]] – PiCyaY – Psychromonas ingrahamii<br /> | ||
| + | **[[4lk8]] – PiCyaY + Co <br /> | ||
| + | **[[4lp1]] – PiCyaY + Eu<br /> | ||
| + | }} | ||
== References == | == References == | ||
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006. | ||
Revision as of 11:41, 15 January 2019
| |||||||||||
3D Structures of frataxin
Updated on 15-January-2019
References
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006.
Proteopedia Page Contributors and Editors (what is this?)
João Victor Paccini Coutinho, Michal Harel, Rebeca B. Candia