We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Frataxin
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
'''General Aspects''' | '''General Aspects''' | ||
| - | '''Frataxin''' is a protein capable of storing, releasing and detoxifying intracellular iron. In humans, a mutation in this protein can trigger the Friedreich's ataxia, a neurodegenerative disease caused due to incapacity to form iron-sulfur groups necessary to activate the mitochondrial enzyme involved in the electron transportation chain, aconitase. | + | '''Frataxin''' (FXN) is a protein capable of storing, releasing and detoxifying intracellular iron. In humans, a mutation in this protein can trigger the Friedreich's ataxia, a neurodegenerative disease caused due to incapacity to form iron-sulfur groups necessary to activate the mitochondrial enzyme involved in the electron transportation chain, aconitase. |
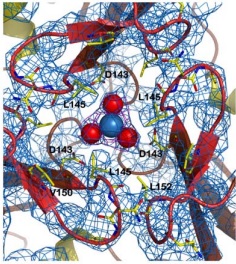
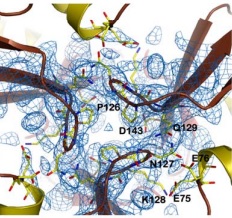
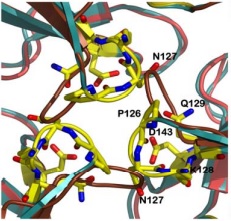
| - | It consists of a polymeric molecule that, | + | It consists of a polymeric molecule that, although capable of forming larger complexes (as the 24 subunit oligomer detected by electron microscopy), exerts its activity by association of three subunits, enough to form a central channel where the ferroxidation takes place. |
| - | In the following paragraphs, we describe the general features of its structure in the trimeric form as | + | In the following paragraphs, we describe the general features of its structure in the trimeric form as obtained by X-ray crystallography at 3Å resolution. The protein used was obtained from the Y37A yeast, which has a 40% sequence identity to the human frataxin. |
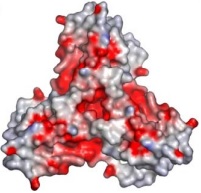
In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | In the box at the right, it is possible to see its <scene name='78/788815/Spacefill_model/1'>general structure</scene> in a space-fill model, in which <font color='violet'><b>violet</b></font>, <font color='orangered'><b>orange</b></font> and <span style="color:aquamarine;background-color:darkgrey;font-weight:bold;">light-green</span> represent, each, a different monomer from the entire molecule. | ||
| Line 18: | Line 18: | ||
'''General Secondary and Tertiary Structure Patterns''' | '''General Secondary and Tertiary Structure Patterns''' | ||
| - | The <scene name='78/788815/Alpha-helices_and_beta-sheets/1'>secondary structure</scene> is basically composed of two alpha-helices (in <font color='red'><b>red</b></font>) and a short helical segment (in <font color='magenta'><b>magenta</b></font>), and seven antiparallel | + | The <scene name='78/788815/Alpha-helices_and_beta-sheets/1'>secondary structure</scene> is basically composed of two alpha-helices (in <font color='red'><b>red</b></font>) and a short helical segment (in <font color='magenta'><b>magenta</b></font>), and seven antiparallel β-sheets, '''in each monomer''', represented here as six <span style="color:springgreen;background-color:darkgrey;font-weight:bold;">green setae</span> and one short region colored <span style="color:yellow;background-color:darkgrey;font-weight:bold;">yellow</span>. <font color='dodgerblue'><b>Turns</b></font> and <span style="color:white;background-color:darkseagreen;font-weight:bold;">coils</span> are also represented. |
You can also view the monomer isolated <scene name='78/788815/Isolated_monomer/1'>here</scene>. | You can also view the monomer isolated <scene name='78/788815/Isolated_monomer/1'>here</scene>. | ||
Revision as of 11:43, 15 January 2019
| |||||||||||
3D Structures of frataxin
Updated on 15-January-2019
References
KARLBERG, Tobias et al. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure, v. 14, n. 10, p. 1535-1546, 2006.
Proteopedia Page Contributors and Editors (what is this?)
João Victor Paccini Coutinho, Michal Harel, Rebeca B. Candia