Introduction
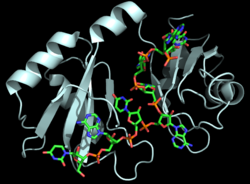
Figure 1: Cartoon representation of the Hrp1-PEE complex. The RNA is shown as a stick model and is colored by element.
Nuclear polyadenylated RNA-binding proteins (NAB) are yeast proteins which may be required for packaging pre-mRNAs into ribonucleoprotein structures amenable to efficient nuclear RNA processing[1].
- Nab2 is essential for cell viability[2].
- Nab3 interacts with the nascent RNA transcript and RNA polymerase II[3].
- Hrp1 or Nab4 is a polyadenylation factor found in Saccharomyces cerevisiae (yeast) [4]. Hrp1 specifically recognizes and binds to an RNA sequence in the 3'UTR of the messenger RNA (mRNA) upstream from the cleavage site called the polyadenylation enhancement element (PEE) (Figure 1) [4]. Upon binding to the RNA, Hrp1 helps recruit additional proteins necessary for the cleavage and polyadenylation of the RNA molecule [4]. Although Hrp1 shares several common features with other RNA-binding proteins, the unique structural features of the Hrp1-PEE complex reveals the mechanism by which Hrp1 is able to recognize and bind to its specific RNA sequence at the atomic level [4]. Hrp1 was discovered when Cleavage Factor I (CF I) was purified and separated into its two components, CF IA and CF IB. CF IB is a single 73 kDa polypeptide. The polypeptide was digested and two tryptic peptides were obtained for sequencing. The sequences were aligned via a database, and Hrp1 was determined to be a perfect match. Hrp1 of CF IB interacts with Rna14 and Rna15 of CF IA[5] to form a protein complex that aids in cleavage and polyadenylation of pre-mRNA and transport of mature mRNA from the nucleus[6].
Structure
General Features
Hrp1 is a single-stranded RNA-binding protein composed of two RNP-type RNA-binding domains (RBDs) arranged in tandem with a typical ßαßßαß architecture [4]. The two RBDs have similar topolgies, both containing a central antiparallel four-stranded with two α-helices running across one face [4]. The β-strands of each βαβ domain are linked via hydrogen bonding between conserved residues, . The two RBDs associate to form a deep and positively charged , which constitutes the binding site for the RNA molecule [4].
Hrp1-RNA Interactions
The interface between Hrp1 and its target RNA sequence is dominated by interactions between key aromatic residues and RNA nucleobases [4]. Only six RNA bases, an repeat, act as the PEE and form specific contacts with Hrp1 [4]. The kinked conformation around Ade4 is uncommon for RNA alone, and may be adopted by the RNA for specific interactions with Hrp1. Ade4 is part of a crucial interaction with Trp168 which will be discussed later, and could explain the adoption of the kinked conformation. Hydrophilic residues of Hrp1 provide base specificity through hydrogen bonding [4]. Most of the key residues that interact with the RNA can be found in the ß-sheet region of Hrp1; however, loops and the interdomain linker are also essential for Hrp1-RNA recognition [4]. Perhaps the most important Hrp1-RNA interaction is the [4]. In this case, Trp168 stacks on Ade4 and forms crucial base-specific hydrogen bonds such as the H-bond between N7 of Ade4 and amide hydrogren of Trp168 [4]. This interaction is a unique feature of the Hrp1-PEE complex and has not been found in any other single-stranded RNA-binding proteins with two canonical RBDs [4]. It is also worth noting that a second Hrp1 residue is critical to holding Ade4 in place, , which stabilizes Ade4 likely via a cation–π interaction. As displayed in the structure, the lysine cationic nitrogen is approximately 4Å away from the pi system, which is within the 6Å range associated with cation-pi interactions. A third contributor, , also stacks with Ura7 to aid in RNA recognition and binding [4].
RBD-RBD Interactions and the Linker Region
As mentioned above, Hrp1 is composed of two RBDs. The RBDs are connected by a (a short two-turn α-helix), which also contains a crucial residue for RNA binding. Ile234 in the linker region holds Ade6 in place in order to ensure proper with the nearby Phe162. Experimental evidence from protein nuclear magnetic resonance (NMR) data [4] suggests that the two RBDs move independently prior to binding the PEE. Upon binding the PEE, the linker region adopts a short helical structure to rigidly hold the RBDs in place relative to each other. Aside from the linker helix, the only other interaction between the RBDs is between Lys231 and Asp271 [4].
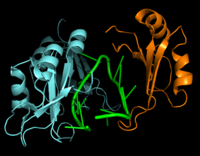
Figure 2: Interaction between Hrp1 (blue), RNA15 (orange) and RNA (green).
Interaction with RNA15
RNA15 is another RNA-binding protein with a single N-terminal RNA recognition motif (RRM) [5]. RNA15 recognizes an A-rich positioning element (PE) downstream from the PEE but upstream from the 3' cleavage site (Figure 2) [5]. The recognition of the PE by RNA15 is crucial for precise cleavage of the RNA molecule. Hrp1 and RNA15 are held together by a separate protein, RNA14 [5]. These proteins act together to anchor the polyadenylation and cleavage protein machinery relative to the cleavage site for precise 3'-end processing [5].
Relationship to other proteins
The RNP-type RBD is found in many proteins involved in post-transcriptional
pre-mRNA processing (5'-end capping, splicing, 3'-end cleavage and polyadenylation, and transport from the nucleus)
[7]. The unique RBD of Hrp1 enables the protein to bind an RNA sequence that differs in both length and content from the RNA sequences of other RNA-binding and mRNA processing proteins such as
sex lethal,
Poly (A)-binding protein (PABP), and
HuD [4]. These proteins, in addition to Hrp1 and
SRp20, contain conserved hydrophobic residues which contribute to hydrophobic interactions between secondary structures of the proteins. Each protein also contains conserved residues L166 and G201 which form a hydrogen bond, linking the β-sheets in the βαβ complex of Hrp1 (Figure 3).
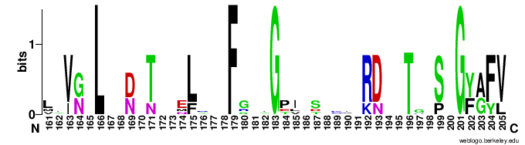
Figure 3: Sequence logo for residues 161-205 of Hrp1.
Like Hrp1, each of these proteins belongs to the class of single-stranded proteins composed of two canonical RBDs; however, each protein is differentiated by respective target RNA sequences, interactions with RNA at the atomic level, and interdomain contacts
[4]. Hrp1 is unique in that HuD, sex lethal, and PABP all contain at least one intra-RNA base-base stacking interaction, a feature that is lacking in the Hrp1-PEE complex
[4]. It is possible that the intra-RNA interactions found in these other protein-RNA complexes is replaced by the crucial Trp168-Ade4 stacking interaction found in the Hrp1 complex
[4]. This may help explain why the Hrp1-RNA interface involves only 6 nucleotides whereas PABP, sex lethal, and HuD require a longer 8-10 nucleotide sequence in the RNA binding pocket
[4].



