We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Retroviral Integrase
From Proteopedia
(Difference between revisions)
| Line 85: | Line 85: | ||
<br /> | <br /> | ||
| + | ==3D structures of retroviral integrase== | ||
| + | [[Retroviral integrase 3D structures]] | ||
| + | |||
| + | </StructureSection> | ||
== 3D Structures of Retroviral Integrase == | == 3D Structures of Retroviral Integrase == | ||
| Line 102: | Line 106: | ||
*Human immunodeficiency virus 1 integrase | *Human immunodeficiency virus 1 integrase | ||
| - | **[[5oi5]], [[5oi3]], [[5oi2]], [[1itg]] – IN catalytic core domain <br /> | + | **[[5oi5]], [[5oi3]], [[5oi2]], [[1itg]], [[6jcg]], [[6jcf]] – IN catalytic core domain <br /> |
**[[3lpt]], [[3lpu]], [[3l3u]], [[1exq]], [[1b92]], [[1b9d]], [[1b9f]], [[1bi4]], [[1bhl]], [[1bl3]], [[1bis]], [[1biu]], [[1biz]], [[2itg]], [[3zcm]], [[5jl4]] - IN catalytic core domain (mutant) – HIV-1<br /> | **[[3lpt]], [[3lpu]], [[3l3u]], [[1exq]], [[1b92]], [[1b9d]], [[1b9f]], [[1bi4]], [[1bhl]], [[1bl3]], [[1bis]], [[1biu]], [[1biz]], [[2itg]], [[3zcm]], [[5jl4]] - IN catalytic core domain (mutant) – HIV-1<br /> | ||
**[[3l3v ]]- IN catalytic core domain (mutant) + sucrose<br /> | **[[3l3v ]]- IN catalytic core domain (mutant) + sucrose<br /> | ||
| Line 114: | Line 118: | ||
**[[2b4j]], [[3av9]], [[3ava]], [[3avb]], [[3avc]], [[3avf]], [[3avg]], [[3avh]], [[3avi]], [[3avj]], [[3avk]], [[3avl]], [[3avm]], [[3avn]] - IN (mutant) + lens epithelium-derived growth factor <br /> | **[[2b4j]], [[3av9]], [[3ava]], [[3avb]], [[3avc]], [[3avf]], [[3avg]], [[3avh]], [[3avi]], [[3avj]], [[3avk]], [[3avl]], [[3avm]], [[3avn]] - IN (mutant) + lens epithelium-derived growth factor <br /> | ||
**[[6ex9]], [[5krt]], [[5krs]], [[5kgx]], [[5kgw]], [[4o0j]] – IN catalytic core domain + inhibitor <br /> | **[[6ex9]], [[5krt]], [[5krs]], [[5kgx]], [[5kgw]], [[4o0j]] – IN catalytic core domain + inhibitor <br /> | ||
| - | **[[3nf6]], [[3nf7]], [[3nf8]], [[3nf9]], [[3nfa]], [[4e1m]], [[4e1n]], [[4cj4]], [[4jlh]], [[4ojr]], [[5hrs]], [[5hrr]], [[5hrp]], [[5hrn]], [[5hot]], [[4tsx]], [[4ovl]], [[4ojr]], [[4o5b]], [[4o55]], [[4nyf]] - IN core domain (mutant) + inhibitor<br /> | + | **[[3nf6]], [[3nf7]], [[3nf8]], [[3nf9]], [[3nfa]], [[4e1m]], [[4e1n]], [[4cj4]], [[4jlh]], [[4ojr]], [[5hrs]], [[5hrr]], [[5hrp]], [[5hrn]], [[5hot]], [[4tsx]], [[4ovl]], [[4ojr]], [[4o5b]], [[4o55]], [[4nyf]], [[6nuj]], [[6ncj]], [[6eb2]], [[6eb1]] - IN core domain (mutant) + inhibitor<br /> |
**[[5tc2]] – IN C terminal <br /> | **[[5tc2]] – IN C terminal <br /> | ||
**[[5eu7]] – IN catalytic core domain + antibody <br /> | **[[5eu7]] – IN catalytic core domain + antibody <br /> | ||
| Line 185: | Line 189: | ||
**[[1ae9]] – λIN catalytic core<br /> | **[[1ae9]] – λIN catalytic core<br /> | ||
**[[5j0n]] – λIN + ATTP + ATTB + integration host factor + excisionase - Cryo EM<br /> | **[[5j0n]] – λIN + ATTP + ATTB + integration host factor + excisionase - Cryo EM<br /> | ||
| + | **[[5udo]] – IN coiled-coil domain – Listeria innocua phage <br /> | ||
*Yeast integrase | *Yeast integrase | ||
Revision as of 08:47, 15 December 2019
| |||||||||||
Integrase Inhibitors
| Name | Brand | Company | Patent | Notes |
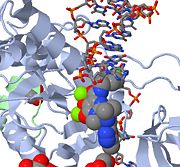
| Raltegravir | Isentress | Merck & Co. | - | also known as MK-0518. The isopropyl and methyl-oxadiazole of MK-0518 are involved in hydrophobic and stacking interactions with side chains of Pro 214 and Tyr 212 to stabilize this drug within the PFV intasome active site. This manner of drug-binding interaction causes displacement of the reactive 3' viral DNA end from the active site of PFV intasome. After binding of MK-0518 to active site, the reactive 3' hydroxyl group moves away from the active site of the PFV intasome by more than 6 Angstroms. Raltegravir was approved by the FDA on October 12, 2007, for use with other anti-HIV agents in the treatment of HIV infection in adults. It is the first integrase inhibitor approved by the FDA. |
| Elvitegravir | - | Gilead Science | - | GS-9137 interacts with Pro 214 of PFV intasome through its quinolone base and isopropyl group. In experimental stages; shares the core structure of quinolone antibiotics. Phase II studies of elvitegravir in people who are treatment experienced have been completed. Phase III studies in treatment experienced patients are ongoing. A phase II study of elvitegravir in people who have never taken antiretroviral therapy is underway. This study will also be evaluated a boosting agent in place of Norvir, currently called GS9350. Elvitegravir holds promise for HIV-positive patients who have taken other anti-HIV drugs in the past. |
| MK-2048 | - | Merck & Co. | - | A second generation integrase inhibitor, intended to be used against HIV infection. It is superior to the first available integrase inhibitor, raltegravir, in that it inhibits the HIV enzyme integrase 4 times longer. It is being investigated for use as part of pre-exposure prophylaxis (PrEP). |
See also Retroviral Integrase Inhibitor Pharmacokinetics[6].
Additional Resources
For additional information, see: Human Immunodeficiency Virus
3D structures of retroviral integrase
Retroviral integrase 3D structures
</StructureSection>
3D Structures of Retroviral Integrase
Updated on 15-December-2019
References
- ↑ Pandey KK, Grandgenett DP. HIV-1 Integrase Strand Transfer Inhibitors: Novel Insights into their Mechanism of Action. Retrovirology (Auckl). 2008 Nov 5;2:11-16. PMID:19915684
- ↑ 2.0 2.1 2.2 2.3 Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010 Mar 11;464(7286):232-6. Epub 2010 Jan 31. PMID:20118915 doi:10.1038/nature08784
- ↑ deJesus, Edwin HIV Antiretroviral Agents in Development. The Body: The Complete HIV/AIDS Resource. March 30, 2006
- ↑ Braun, J.F., Cronje, R.J and Henderson, M.G. HIV-1 Integrase Inhibitors Inhibitors (2008). www.prn.org Volume 13, Pages 1–9
- ↑ AIDS-Info
- ↑ Iwamoto M, Wenning LA, Petry AS, Laethem M, De Smet M, Kost JT, Breidinger SA, Mangin EC, Azrolan N, Greenberg HE, Haazen W, Stone JA, Gottesdiener KM, Wagner JA. Minimal effects of ritonavir and efavirenz on the pharmacokinetics of raltegravir. Antimicrob Agents Chemother. 2008 Dec;52(12):4338-43. Epub 2008 Oct 6. PMID:18838589 doi:10.1128/AAC.01543-07
Further Reading
- GEN News Highlights "Scientists Solve 3-D Crystal Structure of Retroviral Integrase Bound to Viral DNA", Genetic Engineering & Biotechnology News February 1, 2010.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Rhysly Martinez, Joel L. Sussman, Alexander Berchansky, David Canner, Jordan Heard, Eugene Babcock, Garrett Asanuma