We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Amylase
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
α-Amylase is used extensively in various industrial processes. In textile weaving, starch is added for warping. After weaving, the starch is removed by ''Bacillus subtilis'' α-amylase<ref name="book"/>. Dextrin, which is a viscosity improver, filler, or ingredient of food, is manufactured by the liquefaction of starch by bacteria α-amylase<ref name="book"/>. Bacterial α-amylases of ''B.subtilis'', or ''B.licheniformis'' are used for the initial starch liquefaction in producing high conversion glucose syrup<ref name="book"/>. Pancreatitis can be tested by determining the level of amylases in the blood, a result of damaged amylase-producing cells, or excretion due to renal failure<ref>PMID: 16286272 </ref>. α-Amylase is used for the production of malt, as the enzyme is produced during the germination of cereal grains<ref name="book"/>. | α-Amylase is used extensively in various industrial processes. In textile weaving, starch is added for warping. After weaving, the starch is removed by ''Bacillus subtilis'' α-amylase<ref name="book"/>. Dextrin, which is a viscosity improver, filler, or ingredient of food, is manufactured by the liquefaction of starch by bacteria α-amylase<ref name="book"/>. Bacterial α-amylases of ''B.subtilis'', or ''B.licheniformis'' are used for the initial starch liquefaction in producing high conversion glucose syrup<ref name="book"/>. Pancreatitis can be tested by determining the level of amylases in the blood, a result of damaged amylase-producing cells, or excretion due to renal failure<ref>PMID: 16286272 </ref>. α-Amylase is used for the production of malt, as the enzyme is produced during the germination of cereal grains<ref name="book"/>. | ||
β/α amylase (BAAM) is a precursor protein which is cleaved to form the β-amylase and α-amylase after secretion. | β/α amylase (BAAM) is a precursor protein which is cleaved to form the β-amylase and α-amylase after secretion. | ||
| + | |||
| + | = Structure of the AmyC GH13 alpha-amylase from Alicyclobacillus sp, reveals accommodation of starch branching points in the alpha-amylase family = | ||
| + | <big>Jon Agirre, Olga Moroz, Sebastian Meier, Jesper Brask, Astrid Munch, Tine Hoff, Carsten Andersen, Keith S. Wilsona and Gideon J. Davies</big> <ref>doi 10.1107/S2059798318014900</ref> | ||
| + | <hr/> | ||
| + | <b>Molecular Tour</b><br> | ||
| + | The enzymatic degradation of starch has a myriad industrial applications. However, the branched nature of the polysaccharides that compose it poses problems, as branches have to be accommodated within an active centre best suited to linear polysaccharides. Alpha-amylases are glycoside hydrolases that break the α-1,4 bonds in starch and related glycans. The present work provides a rare insight into branch-point acceptance in these industrial catalysts. | ||
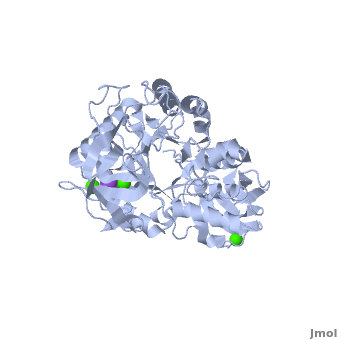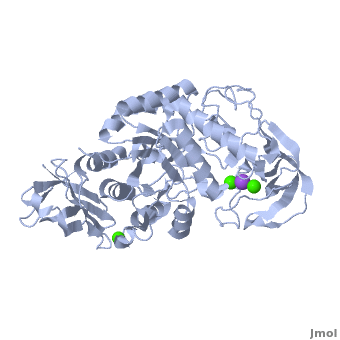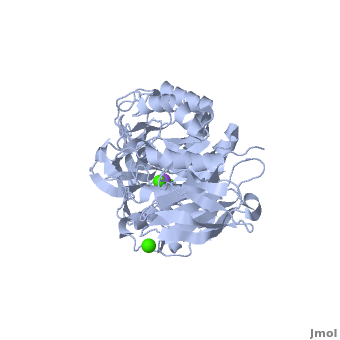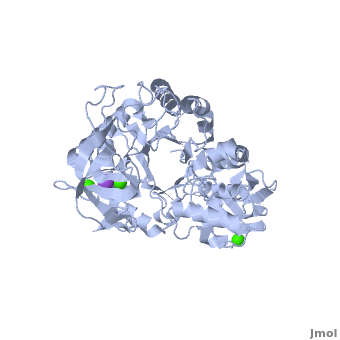
| + | |||
| + | The complex of α-amylase from ''Alicyclobacillus sp.'' 18711 (AliC) with acarbose was solved by molecular replacement, with two molecules of AliC in the asymmetric unit, at a resolution of 2.1 Å ([[6gxv]]). The fold, as expected, is a canonical <scene name='79/799580/Cv1/5'>three-domain arrangement</scene> with the A, B and C domains defined approximately as <span style="color:deepskyblue;background-color:black;font-weight:bold;">A, residues 4–104 and 210–397 (in deepskyblue)</span>, <span style="color:yellow;background-color:black;font-weight:bold;">B, residues 105–209 (in yellow)</span>, and <span style="color:white;background-color:black;font-weight:bold;">C, residues 398–484 (in white)</span>. A classical Ca<sup>2+</sup>–Na<sup>+</sup>–Ca<sup>2+</sup> <scene name='79/799580/Cv1/4'>triad</scene> <ref name="Machius">PMID:9551551</ref>,<ref name="Brzozowski">PMID:10924103</ref> is found at the A/B-domain interface. The structure of AliC was determined in the presence of the <scene name='79/799580/Cv1/6'>inhibitor acarbose</scene> (<span style="color:lime;background-color:black;font-weight:bold;">colored in green</span>). As with many (retaining) α-amylase complexes, the acarbose is observed as a transglycosylated species, here a hexasaccharide which contains two of the acarviosin disaccharide motifs. The <scene name='79/799580/Cv/11'>complex defines six subsites</scene>, -4 to +2, with the expected catalytic GH13 signature triad of Asp234 (nucleophile), Glu265 (acid/base) and <scene name='79/799580/Cv/13'>Asp332 (interacting with O2/O3 of the -1 subsite sugar)</scene> all disposed for catalysis, here around the <sup>2</sup>H<sub>3</sub> half-chair of the unsaturated cyclohexitol moiety. AliC must also be able to accommodate branching in the +2 subsite, which is consistent with the <scene name='79/799580/Cv/14'>glucose moiety seen adjacent to O6 of the +2 sugar</scene>. | ||
| + | *<scene name='79/799580/Cv/12'>Asp234 and Glu265 interactions</scene>. | ||
| + | |||
| + | A ‘branched-ligand’ AliC complex was obtained through co-crystallization, with crystals forming in a new space group. This form diffracted poorly and data could only be obtained to 2.95 Å resolution [[6gya]]). Weak density in the -1 subsite, largely diffuse but greater than would be expected for discrete solvent, remained unmodelled. Density was clearer for a panose trisaccharide with an α-1,4-linked disaccharide in subsites +1 and +2 and, crucially, clear density for an α-1,6 branch accommodated in the +1 subsite, providing a structural context for the limit digest analysis of action on amylopectin starch. The <scene name='79/799580/Cv/15'>binding of the branched oligosaccharide in subsites +1, +2 and +1'</scene> (<span style="color:lime;background-color:black;font-weight:bold;">oligosaccharide colored in green</span>). | ||
=3D structures of amylase= | =3D structures of amylase= | ||
Revision as of 12:00, 28 May 2019
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press
- ↑ 2.0 2.1 Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998 Mar;7(3):564-72. PMID:9541387
- ↑ 3.0 3.1 3.2 Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability. J Biochem. 2001 Mar;129(3):461-8. PMID:11226887
- ↑ French D. Amylases: enzymatic mechanisms. Basic Life Sci. 1981;18:151-82. PMID:6168260
- ↑ Hii SL, Tan JS, Ling TC, Ariff AB. Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012;2012:921362. doi: 10.1155/2012/921362. Epub 2012 Sep 6. PMID:22991654 doi:http://dx.doi.org/10.1155/2012/921362
- ↑ 6.0 6.1 6.2 Aghajari N, Feller G, Gerday C, Haser R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002 Jun;11(6):1435-41. PMID:12021442
- ↑ Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 8.0 8.1 Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 9.0 9.1 9.2 9.3 Kuriki T, Imanaka T. The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87(5):557-65. PMID:16232518
- ↑ 10.0 10.1 PPMID: 17713601
- ↑ Franco OL, Rigden DJ, Melo FR, Grossi-De-Sa MF. Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases. Eur J Biochem. 2002 Jan;269(2):397-412. PMID:11856298
- ↑ Yang RW, Shao ZX, Chen YY, Yin Z, Wang WJ. Lipase and pancreatic amylase activities in diagnosis of acute pancreatitis in patients with hyperamylasemia. Hepatobiliary Pancreat Dis Int. 2005 Nov;4(4):600-3. PMID:16286272
- ↑ Agirre J, Moroz O, Meier S, Brask J, Munch A, Hoff T, Andersen C, Wilson KS, Davies GJ. The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family. Acta Crystallogr D Struct Biol. 2019 Jan 1;75(Pt 1):1-7. doi:, 10.1107/S2059798318014900. Epub 2019 Jan 4. PMID:30644839 doi:http://dx.doi.org/10.1107/S2059798318014900
- ↑ Machius M, Declerck N, Huber R, Wiegand G. Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad. Structure. 1998 Mar 15;6(3):281-92. PMID:9551551
- ↑ Brzozowski AM, Lawson DM, Turkenburg JP, Bisgaard-Frantzen H, Svendsen A, Borchert TV, Dauter Z, Wilson KS, Davies GJ. Structural analysis of a chimeric bacterial alpha-amylase. High-resolution analysis of native and ligand complexes. Biochemistry. 2000 Aug 8;39(31):9099-107. PMID:10924103
Proteopedia Page Contributors and Editors (what is this?)
Shane Riley, Michal Harel, Joel L. Sussman, Randi Woodbeck, Jaime Prilusky, Alexander Berchansky, Ann Taylor, Andrea Gorrell, David Canner