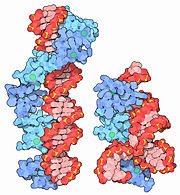
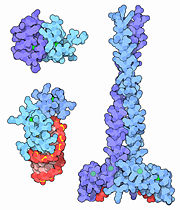
Zinc Fingers
From Proteopedia
| Line 10: | Line 10: | ||
== Basic Structural Profile of a Zinc Finger == | == Basic Structural Profile of a Zinc Finger == | ||
| - | The simplest group of zinc fingers, referred as [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>] zinc fingers, consists of <scene name='10/100440/2ndary_structure/1'>two antiparallel β-pleated sheets and a right-handed α-helix</scene>. The name [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>], or Cis<sub>2</sub>His<sub>2</sub>, gives a nod to the <scene name='10/100440/His_cys_zn/1'>two cysteine and two histidine</scene> residues involved in coordination of the zinc ion. The turn between the two β-pleated sheets forms a hydrophobic pocket near where the zinc ion is bound. Typically, the hydrophobic pocket is formed as a result of interactions between <scene name='10/100440/His_cys_zn_phe/1'>phenylalanines</scene> and leucines in close proximity. While both the coordination of the zinc ligand and the presence of the hydrophobic pocket stabilize the small zinc finger domain, the zinc ligand is responsible for a majority of the stability imparted to the motif. Because cells contain a highly reducing environment, sulfide bridges are unable to stabilize small protein domains. With a single oxidation state and the ability to accommodate both nitrogen and sulfur, Zinc is an ideal stabilizer. Due to the structural stability imparted to the motif by the zinc ion, zinc fingers are considerably smaller than most other proteins, usually ranging between 25 and 30 amino acids in length. As a corollary of their small size, they are extremely agile and mobile, and so are the genes that encode them. Thus, they are easily able to bind to DNA, among other substrates. By slipping into the major groove of DNA, they are able to use their amino acids to check for proper base identity. This allows the zinc fingers to bind to sequences normally unavailable to other, larger DNA-binding structural motifs. | + | The simplest group of zinc fingers, referred as [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>] zinc fingers, consists of <scene name='10/100440/2ndary_structure/1'>two antiparallel β-pleated sheets and a right-handed α-helix</scene>. The name [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>], or Cis<sub>2</sub>His<sub>2</sub>, gives a nod to the <scene name='10/100440/His_cys_zn/1'>two cysteine and two histidine</scene> residues involved in coordination of the zinc ion. The turn between the two β-pleated sheets forms a hydrophobic pocket near where the zinc ion is bound. Typically, the hydrophobic pocket is formed as a result of interactions between <scene name='10/100440/His_cys_zn_phe/1'>phenylalanines</scene> and leucines in close proximity. While both the coordination of the zinc ligand and the presence of the hydrophobic pocket stabilize the small zinc finger domain, the zinc ligand is responsible for a majority of the stability imparted to the motif. Because cells contain a highly reducing environment, sulfide bridges are unable to stabilize small protein domains. With a single oxidation state and the ability to accommodate both nitrogen and sulfur, Zinc is an ideal stabilizer. Due to the structural stability imparted to the motif by the zinc ion, zinc fingers are considerably smaller than most other proteins, usually ranging between 25 and 30 amino acids in length. |
| + | As a corollary of their small size, they are extremely agile and mobile, and so are the genes that encode them. Thus, they are easily able to bind to DNA, among other substrates. By slipping into the major groove of DNA, they are able to use their <scene name='10/100440/Dna-protein_interaction/1'>amino acids</scene> to check for proper base identity. This allows the zinc fingers to bind to sequences normally unavailable to other, larger DNA-binding structural motifs. | ||
| + | |||
| + | As a family, the structure of the zinc fingers is as polymorphous as it is unique. Some are coordinated primarily or exclusively by cysteine, and many form much more complex structures than the β-hairpin of Cis<sub>2</sub>His<sub>2</sub> types. | ||
== Biological Role and Regulation == | == Biological Role and Regulation == | ||
Revision as of 03:53, 1 April 2019
| |||||||||||
3D Structure of Zinc Finger Domains
1hvo - HIV-1 nucleocpasid
1x68 - FHL5
1x6a - LIM kinase 2
1x6f - Zinc finger protein 462
2bai - Genome polyprotein
2ctu - Zinc finger protein 483
2ds6 - Clp Protease
2e72 - Pogo transposable element
2epc - Zinc finger protein 32
2epp - Zinc finger protein 278
2epq - Zinc finger protein 278
2epr - Zinc finger protein 278
2eps - Zinc finger protein 278
2ept - Zinc finger protein 32
2epu - Zinc finger protein 32
2epv - Zinc finger protein 268
2epw - Zinc finger protein 268
2epx - Zinc finger protein 28
2epy - Zinc finger protein 268
2epz - Zinc finger protein 28
2eq0 - Zinc finger protein 347
2eq1 - Zinc finger protein 347
2eq2 - Zinc finger protein 347
2eq3 - Zinc finger protein 347
2eq4 - Zinc finger protein 224
2rpp - Muscleblind-like protein 2
2ysv - Zinc finger protein 473
Additional Information
- Zinc Finger in Wikipedia
- Zinc Finger Consortium
- Molecule of the Month on Zinc Fingers by David Goodsell
- Molecule of the Month at Teaching Scenes, Tutorials, and Educators' Pages
- For Additional Information, See: Transcription
Content Donators
Many images on this page are the work of David S. Goodsell, who has given permission for their inclusion in Proteopedia:
- Content adapted with permission from David S. Goodsell's Molecule of the Month on Zinc Fingers
Proteopedia Page Contributors and Editors (what is this?)
Ala Jelani, Ann Taylor, Eran Hodis, Michal Harel, Tyler Combs, Joel L. Sussman, David Canner, Eric Martz