User:Asif Hossain/Sandbox 1
From Proteopedia
| Line 18: | Line 18: | ||
===Key Residues=== | ===Key Residues=== | ||
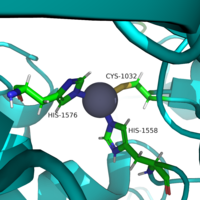
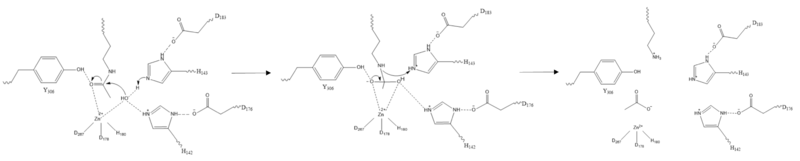
| - | The <scene name='81/811085/Active_site/6'>active site</scene> of HDAC8 is composed of 2 catalytic dyads: | + | The <scene name='81/811085/Active_site/6'>active site</scene> of HDAC8 is composed of 2 catalytic dyads: <scene name='81/811085/Dyads/2'>His143/Asp183 and His142/Asp176</scene>, which activate the catalytic water nucleophile (Figure 2). A Tyr306, through mutation to Phe in the pdb file 2v5w (modeled in the overall view) was observed to render the protein mostly inactive. Thus, it has been hypothesized that the this residue is critical for stabilization of the transition state with the Zn<sup>2+</sup> ion. |
[[Image:Triad.png |200px|left|thumb|Figure 2: Similar to many serine and zinc proteases, HDAC8 uses a mechanism with a "catalytic triad." Instead of the Asp-His-Ser, HDAC8 uses the His to coordinate a H<sub>2</sub>O nucleophile.]] | [[Image:Triad.png |200px|left|thumb|Figure 2: Similar to many serine and zinc proteases, HDAC8 uses a mechanism with a "catalytic triad." Instead of the Asp-His-Ser, HDAC8 uses the His to coordinate a H<sub>2</sub>O nucleophile.]] | ||
===Binding Pocket=== | ===Binding Pocket=== | ||
| - | By encasing, the nonpolar 4 carbon-long side chain of the Lys residue on the | + | By encasing, the nonpolar 4 carbon-long side chain of the Lys residue on the ligand, Phe152 and Phe208 engage in hydrophobic Van der Waals with the ligand. & At different ends of the <scene name='81/811085/Binding_site/2'>Binding Pocket</scene>, W141 and M274 contribute to the overall shape through general hydrophobic interactions.<ref name="Whitehead">Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030 </ref> Finally, the carbonyl O of Gly151 hydrogen bonds with the amide H of the acetylated lysine to further interact with the ligand in the relatively hydrophobic tunnel.** |
Revision as of 21:22, 8 April 2019
Contents |
Histone Deacetylase 8 (HDAC 8)
Introduction
Histone deacetylase 8 (HDAC8) is an enzyme that plays a role in controlling gene expression in Homo sapiens. Specifically, HDAC8 catalyzes the removal of an acetyl group off of the ε-amino-lysine sidechain of N-terminal core of histones. (NEED REFERENCE HERE) Histones consist of eight monomers to form an octomer complex. In addition, histones are highly basic and have a large positive charge. Since DNA is negatively charged, histones tightly interact with DNA. This prevents transcription factors from accessing DNA, thus decreasing gene expression. Chromatin remodeling by histone acetylation and/or deacetylation is an example of epigenetic regulation. Histone acetlytransferase (HAT1) catalyze the addition of an acetyl group onto a histone. The lack of charge of the acetyl group prevents the interaction between DNA and histones. This allows transcription factors to access the DNA to increase gene expression. HDAC8 though reverses this reaction by catalyzing the removal of these acetyl groups by removing the acetate and the reclaimed positive charge on the lysine sidechain is able to interact with the negative charge on the DNA. As a result, DNA will bind more tightly to the histone protein, repressing transcription and gene expression. (NEED REFERENCE HERE)
| |||||||||||
Medical Relevance
Besides controlling the gene regulation through deacetylation of histones, HDAC 8 also regulates the post-transcriptional acetylation status of many non-histone proteins, including transcription factors, chaperones, hormone receptors and signaling molecules. Thus, it has influences on protein stability, protein-protein interactions and protein-DNA interactions. HDAC 8 can therefore affect the regulation of cell proliferation and cell death. These processes are typically being altered in cancer cells and that makes HDAC enzymes an interesting potential target for cancer drugs. HDAC inhibitors have been shown to be promising cancer drug agents in prior research as the HDAC inhibitors cease tumor growth in cancer cells by either making them differentiate, undergo apoptosis or upregulate cell cycle arrest proteins. [4] One way, the HDAC inhibitors ceases tumor growth is by the reactivation of the transcription factor,RUNX3, a known tumor suppressor. HDACi increases the acetylation of the protein and as the stability of RUNX3 is dependent on the acetylation status of the protein, the increased acetylation or HDAC inhibition will enhance the protein stability, causing an increase in the anti-tumorous properties of the protein. A number of HDAC inhibitors have been purified from natural sources or synthesized and at least four structurally different inhibitor classes have been characterized: hydrox-amates, cyclic peptides, aliphatic acids and ben-zamides. The Vorinostat(within the hydroxamate class) has been FDA-approved for treatment of cancer. The hydroxamate HDAC inhibitors consists of a metal-binding domain, a linker domain and a hydrophobic capping group. The HDAC class 1 hydroxamic acid, compound 1, fashion in a bidendate fashion while making hydrogen bonds to important residues as at the active site of HDAC8. Thus, the HDAC inhibitors can be used as antagonists to prevent the functioning of HDAC8 in cancer treatment. [5]
References
- ↑ Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ 2.0 2.1 2.2 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414
- ↑ Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101