User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
===Ser 39=== | ===Ser 39=== | ||
The <scene name='81/811087/Ser39_at_the_surface_of_hdac8/1'>Ser39</scene> has been shown to play a role in the activity of HDAC8 as the HDAC8 activity is regulated by phosphorylation at Ser39 by [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A]. The phosphorylation of Ser39 leads to a decrease in the enzyme's activity. Ser39 lies at the surface of HDAC8, roughly 20 Å from the opening to the HDAC8 active site and it could be forming part of the surface that interacts with the target histone. The phosphorylation of Ser39 could disrupt the interaction between HDAC8 and the target histone. In addition, the phosphorylated Ser39 provokes a structural rearrangement near the active site by <scene name='81/811087/Active_site_loop_1_s30-k36/5'>interacting with structural elements as K36</scene>, part of the conformational active loop L1, that extends into the active site. The Ser39 phosphorylation could therefore be inducing a conformation of the L1 loop that prohibits a competent substrate binding. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | The <scene name='81/811087/Ser39_at_the_surface_of_hdac8/1'>Ser39</scene> has been shown to play a role in the activity of HDAC8 as the HDAC8 activity is regulated by phosphorylation at Ser39 by [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A]. The phosphorylation of Ser39 leads to a decrease in the enzyme's activity. Ser39 lies at the surface of HDAC8, roughly 20 Å from the opening to the HDAC8 active site and it could be forming part of the surface that interacts with the target histone. The phosphorylation of Ser39 could disrupt the interaction between HDAC8 and the target histone. In addition, the phosphorylated Ser39 provokes a structural rearrangement near the active site by <scene name='81/811087/Active_site_loop_1_s30-k36/5'>interacting with structural elements as K36</scene>, part of the conformational active loop L1, that extends into the active site. The Ser39 phosphorylation could therefore be inducing a conformation of the L1 loop that prohibits a competent substrate binding. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | ||
| - | |||
==Mechanism== | ==Mechanism== | ||
| Line 42: | Line 41: | ||
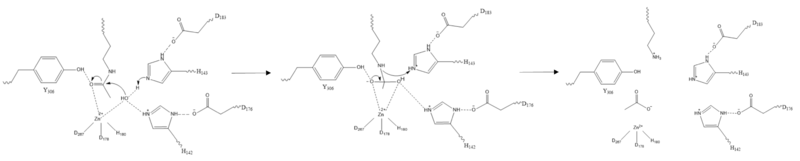
Besides controlling the gene regulation through deacetylation of histones, HDAC 8 also regulates the post-transcriptional acetylation status of many non-histone proteins, including transcription factors, chaperones, hormone receptors and signaling molecules. Thus, it has influences on protein stability, protein-protein interactions and protein-DNA interactions. HDAC 8 can therefore affect the regulation of cell proliferation and cell death. These processes are typically being altered in cancer cells and that makes HDAC enzymes an interesting potential target for cancer drugs. HDAC inhibitors have been shown to be promising cancer drug agents in prior research as the HDAC inhibitors cease tumor growth in cancer cells by either making them differentiate, undergo apoptosis or upregulate cell cycle arrest proteins. <ref name="Eckschlager">Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414</ref> One way, the HDAC inhibitors ceases tumor growth is by the reactivation of the transcription factor, [https://en.wikipedia.org/wiki/RUNX3 RUNX3], a known tumor suppressor. HDACi increases the acetylation of the protein and as the stability of RUNX3 is dependent on the acetylation status of the protein, the increased acetylation or HDAC inhibition will enhance the protein stability, causing an increase in the anti-tumorous properties of the protein. A number of HDAC inhibitors have been purified from natural sources or synthesized and at least four structurally different inhibitor classes have been characterized: hydrox-amates, cyclic peptides, aliphatic acids and ben-zamides. The Vorinostat(within the hydroxamate class) has been FDA-approved for treatment of cancer. The hydroxamate HDAC inhibitors consists of a metal-binding domain, a linker domain and a hydrophobic capping group. The HDAC class 1 hydroxamic acid, compound 1,<scene name='81/811087/Inhibitor_and_zinc_binding/2'> binds to the zinc ion</scene> in a bidendate fashion while making hydrogen bonds to important residues as <scene name='81/811087/Inhibitor_and_zinc_binding/4'>His142, His143, and Tyr306</scene> at the active site of HDAC8. Thus, the HDAC inhibitors can be used as antagonists to prevent the functioning of HDAC8 in cancer treatment. <ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref> | Besides controlling the gene regulation through deacetylation of histones, HDAC 8 also regulates the post-transcriptional acetylation status of many non-histone proteins, including transcription factors, chaperones, hormone receptors and signaling molecules. Thus, it has influences on protein stability, protein-protein interactions and protein-DNA interactions. HDAC 8 can therefore affect the regulation of cell proliferation and cell death. These processes are typically being altered in cancer cells and that makes HDAC enzymes an interesting potential target for cancer drugs. HDAC inhibitors have been shown to be promising cancer drug agents in prior research as the HDAC inhibitors cease tumor growth in cancer cells by either making them differentiate, undergo apoptosis or upregulate cell cycle arrest proteins. <ref name="Eckschlager">Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414</ref> One way, the HDAC inhibitors ceases tumor growth is by the reactivation of the transcription factor, [https://en.wikipedia.org/wiki/RUNX3 RUNX3], a known tumor suppressor. HDACi increases the acetylation of the protein and as the stability of RUNX3 is dependent on the acetylation status of the protein, the increased acetylation or HDAC inhibition will enhance the protein stability, causing an increase in the anti-tumorous properties of the protein. A number of HDAC inhibitors have been purified from natural sources or synthesized and at least four structurally different inhibitor classes have been characterized: hydrox-amates, cyclic peptides, aliphatic acids and ben-zamides. The Vorinostat(within the hydroxamate class) has been FDA-approved for treatment of cancer. The hydroxamate HDAC inhibitors consists of a metal-binding domain, a linker domain and a hydrophobic capping group. The HDAC class 1 hydroxamic acid, compound 1,<scene name='81/811087/Inhibitor_and_zinc_binding/2'> binds to the zinc ion</scene> in a bidendate fashion while making hydrogen bonds to important residues as <scene name='81/811087/Inhibitor_and_zinc_binding/4'>His142, His143, and Tyr306</scene> at the active site of HDAC8. Thus, the HDAC inhibitors can be used as antagonists to prevent the functioning of HDAC8 in cancer treatment. <ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref> | ||
</StructureSection> | </StructureSection> | ||
| - | |||
| - | |||
| - | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 19:05, 9 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414
- ↑ Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101