We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Madeleine Wilson/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
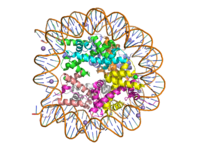
Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. (1) Modifications to histone core proteins can affect the accessibility of genes in the genome and their ability to be transcribed. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. (2) | Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. (1) Modifications to histone core proteins can affect the accessibility of genes in the genome and their ability to be transcribed. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. (2) | ||
| - | Specifically, histone methylation is associated with gene activation. (3) Many domain families fall under the Histone methylase family, one of these enzymes being the SET7 domain family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. (4) Some tumor related genes such as p53 are site specifically methylated to promote biological function (5), whereas hypomethylation of CpG is linked to tumor genesis. (2) A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. (4,3) | + | Specifically, histone methylation is associated with gene activation. (3) Many domain families fall under the Histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_rotate/4'>SET7 domain</scene> family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. (4) Some tumor related genes such as p53 are site specifically methylated to promote biological function (5), whereas hypomethylation of CpG is linked to tumor genesis. (2) A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. (4,3) |
==KMT Structure== | ==KMT Structure== | ||
Revision as of 00:58, 10 April 2019
Lysine Methyl Transferase, Homo Sapiens
| |||||||||||