We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sean Callahan/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
==Application== | ==Application== | ||
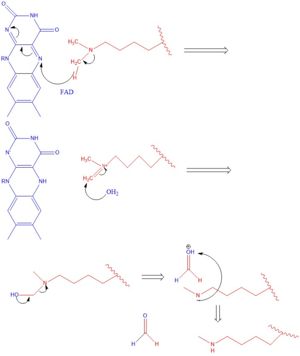
| - | LSD1 plays a pivotal role in many biological processes such as cell growth, epithelial-mesenchymal transition, stem cell biology, malignant transformation of cells, and cell differentiation. Malfunctioning of these activities can lead to life threatening diseases such as acute myeloid leukemia and acute lymphoblastic leukemia. Evidence has shown that these activities have ties to prostate and small cell lung cancer so studies have been done to find a reliable inhibitor for LSD1.[[Image:Formula 1.jpg|350px|right|thumb|Figure 3]] Monamine oxidases utilize the FAD co-factor much like LSD1 and inhibitors of monamine oxidase have proven to be successful in inhibiting the activity of LSD1. Thus inhibition of LSD1 opens new pathways for possible treatments for cancers and disorders <ref name="Abdel-Magid">PMID:29152043</ref>.</StructureSection> | + | LSD1 plays a pivotal role in many biological processes such as cell growth, epithelial-mesenchymal transition, stem cell biology, malignant transformation of cells, and cell differentiation. Malfunctioning of these activities can lead to life threatening diseases such as acute myeloid leukemia and acute lymphoblastic leukemia. Evidence has shown that these activities have ties to prostate and small cell lung cancer so studies have been done to find a reliable inhibitor for LSD1.[[Image:Formula 1.jpg|350px|right|thumb|Figure 3: Derivatives of this formula 1 compound have shown to inhibit LSD1's activity through reversible and irreversible means.]] Monamine oxidases utilize the FAD co-factor much like LSD1 and inhibitors of monamine oxidase have proven to be successful in inhibiting the activity of LSD1. Thus inhibition of LSD1 opens new pathways for possible treatments for cancers and disorders <ref name="Abdel-Magid">PMID:29152043</ref>.</StructureSection> |
== References == | == References == | ||
| - | <ref name=”Qian">PMID: 16299514 </ref> | ||
<ref name=”Forneris">PMID: 16223729 </ref> | <ref name=”Forneris">PMID: 16223729 </ref> | ||
<ref name="Forneris">PMID: 15811342</ref> | <ref name="Forneris">PMID: 15811342</ref> | ||
| Line 42: | Line 41: | ||
<ref name="Stavropoulos">PMID: 16799558</ref> | <ref name="Stavropoulos">PMID: 16799558</ref> | ||
<ref name="Abdel-Magid">PMID:29152043</ref> | <ref name="Abdel-Magid">PMID:29152043</ref> | ||
| + | <ref name=”Qian">PMID: 16299514 </ref> | ||
<references/> | <references/> | ||
Revision as of 18:27, 12 April 2019
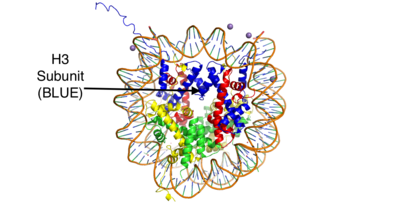
Lysine Specific Demethylase 1 (Homo sapiens)
| |||||||||||
References
- ↑ 1.0 1.1 Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005 Apr 11;579(10):2203-7. doi: 10.1016/j.febslet.2005.03.015. PMID:15811342 doi:http://dx.doi.org/10.1016/j.febslet.2005.03.015
- ↑ Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005 Dec;12(12):1078-85. Epub 2005 Nov 20. PMID:16299514 doi:10.1038/nsmb1022
- ↑ 3.0 3.1 Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2057-62. Epub 2006 Feb 3. PMID:16461455
- ↑ 4.0 4.1 4.2 4.3 4.4 Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006 Jul;13(7):626-32. Epub 2006 Jun 25. PMID:16799558 doi:10.1038/nsmb1113
- ↑ 5.0 5.1 Abdel-Magid AF. Lysine-Specific Demethylase 1 (LSD1) Inhibitors as Potential Treatment for Different Types of Cancers. ACS Med Chem Lett. 2017 Oct 27;8(11):1134-1135. doi:, 10.1021/acsmedchemlett.7b00426. eCollection 2017 Nov 9. PMID:29152043 doi:http://dx.doi.org/10.1021/acsmedchemlett.7b00426
- ↑ Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005 Dec 16;280(50):41360-5. doi: 10.1074/jbc.M509549200. Epub 2005 , Oct 13. PMID:16223729 doi:http://dx.doi.org/10.1074/jbc.M509549200
- ↑ Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005 Dec;12(12):1078-85. Epub 2005 Nov 20. PMID:16299514 doi:10.1038/nsmb1022