User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Histone Acetyltransferase HAT1/HAT2 Complex, ''Saccharomyces cerevisiae''== | ==Histone Acetyltransferase HAT1/HAT2 Complex, ''Saccharomyces cerevisiae''== | ||
<StructureSection load='4psw' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW'> | <StructureSection load='4psw' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW'> | ||
| - | + | = Introduction = | |
| - | =Histones= | + | ==Histones== |
[https://en.wikipedia.org/wiki/Histone Histones]are essential for proper DNA packaging and are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin]. They are subject to post-translational modifications and play important roles in replication, transcription, heterochromatin maintenance, and DNA repair. | [https://en.wikipedia.org/wiki/Histone Histones]are essential for proper DNA packaging and are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin]. They are subject to post-translational modifications and play important roles in replication, transcription, heterochromatin maintenance, and DNA repair. | ||
| - | =Histone Modification= | + | ==Histone Modification== |
Histones can be modified in a variety of ways, including: methylations, demethylation, acetylation, and deacetylation, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification. This involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by the histone acetyltransferase (HAT) enzyme families. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. | Histones can be modified in a variety of ways, including: methylations, demethylation, acetylation, and deacetylation, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification. This involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by the histone acetyltransferase (HAT) enzyme families. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. | ||
| - | == HAT1 Background == | + | ==HAT1 Background == |
<scene name='81/811717/Hat1_with_accoa/1'>HAT1</scene> was the first of the HAT enzymes to be identified in yeast. It is lysine specific for newly synthesized histone 4 (H4). One study showed that the deletion of the HAT caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for the evolutionary conserved histone modification.<ref name="Parthun">PMID:8858151</ref> The HAT2 enzyme is identified as a binding partner for HAT1 to help modulate the substrate specificity of HAT1. The complex is highly specific for H4K12. | <scene name='81/811717/Hat1_with_accoa/1'>HAT1</scene> was the first of the HAT enzymes to be identified in yeast. It is lysine specific for newly synthesized histone 4 (H4). One study showed that the deletion of the HAT caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for the evolutionary conserved histone modification.<ref name="Parthun">PMID:8858151</ref> The HAT2 enzyme is identified as a binding partner for HAT1 to help modulate the substrate specificity of HAT1. The complex is highly specific for H4K12. | ||
| - | + | = Hat1/Hat2 Complex Structure = | |
Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/1'>complex</scene>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/1'>chain A</scene>, includes 317 residues and contains the active site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/1'>chain B</scene>, which includes 401 residues. The activated complex acetylates residues in the 38 residue span of <scene name='81/811717/Histone_4/2'>Histone 4</scene> . | Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/1'>complex</scene>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/1'>chain A</scene>, includes 317 residues and contains the active site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/1'>chain B</scene>, which includes 401 residues. The activated complex acetylates residues in the 38 residue span of <scene name='81/811717/Histone_4/2'>Histone 4</scene> . | ||
| Line 17: | Line 17: | ||
Once the complex has formed, histone 4 and AcetylCoA can begin to interact. The N-terminal segment of H4 that binds with HAT1/HAT2 can be divided into <scene name='81/811717/Ntail_regions/1'>three different regions</scene>. Similar to 1BOB the N-terminal region of H4 is embedded in a cave between HAT1 and HAT2, though H4 mainly interacts with HAT1. The C-terminal helix of H4 is found inserted into LP2, the N-terminal helix, and C-terminal groove of HAT2. These interactions are strongly stabilized by salt bridge bonds between the histone and the complex. Previous studies suggest that H4K12 inserts into the active site of HAT1 to access AcCoA.<ref name="Wu">PMID:22615379</ref> The 4PSW structure has H4K12 aligned with the active site and the AcCoA entering the concave groove from the opposite side. This allows the ε-amino group of H4K12 to contact the SH group of CoA. | Once the complex has formed, histone 4 and AcetylCoA can begin to interact. The N-terminal segment of H4 that binds with HAT1/HAT2 can be divided into <scene name='81/811717/Ntail_regions/1'>three different regions</scene>. Similar to 1BOB the N-terminal region of H4 is embedded in a cave between HAT1 and HAT2, though H4 mainly interacts with HAT1. The C-terminal helix of H4 is found inserted into LP2, the N-terminal helix, and C-terminal groove of HAT2. These interactions are strongly stabilized by salt bridge bonds between the histone and the complex. Previous studies suggest that H4K12 inserts into the active site of HAT1 to access AcCoA.<ref name="Wu">PMID:22615379</ref> The 4PSW structure has H4K12 aligned with the active site and the AcCoA entering the concave groove from the opposite side. This allows the ε-amino group of H4K12 to contact the SH group of CoA. | ||
| - | + | ==Ligand Active Site== | |
The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is <scene name='81/811717/Tight_protein-ligand_intxn/1'>buried in the protein</scene> (~60%). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix alpha 7, C terminal end of strand beta-14/loop Beta15-Alpha9, and N-terminal half of helix alpha 9 <ref name=”Dutnall”>PMID:10384314</ref>. | The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is <scene name='81/811717/Tight_protein-ligand_intxn/1'>buried in the protein</scene> (~60%). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix alpha 7, C terminal end of strand beta-14/loop Beta15-Alpha9, and N-terminal half of helix alpha 9 <ref name=”Dutnall”>PMID:10384314</ref>. | ||
| Line 23: | Line 23: | ||
| - | + | = Mechanism = | |
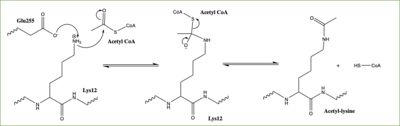
Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. Previous studies found that a mutation at the active site glutamate residue greatly alters the catalytic ability of HAT1, proving it to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structural information from the crystallized structure of the HAT1/HAT2 complex regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactor: <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, Lys14, and AcetylCoA</scene>. | Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. Previous studies found that a mutation at the active site glutamate residue greatly alters the catalytic ability of HAT1, proving it to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structural information from the crystallized structure of the HAT1/HAT2 complex regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactor: <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, Lys14, and AcetylCoA</scene>. | ||
| Line 31: | Line 31: | ||
In this mechanism, the glutamate at residue 255 in the active site of the protein acts as a general base by deprotonating lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues in the pdb file 4psw).The deprotonated lysine then acts as a nucleophile and attacks the carbonyl carbon of acetyl coenzyme Acetyl CoA (not shown in pdb file), forming a tetrahedral transition state with an oxyanion. The negative charge on the oxyanion then shift to down to reform the double bond between the oxygen and carbonyl carbon, breaking the scissle bond between the carbonyl carbon and the sulfur atom of acetyl CoA. The resulting product of this reaction is histone 4 with an acetyl-lysine at residue 12 and CoEnzyme A. | In this mechanism, the glutamate at residue 255 in the active site of the protein acts as a general base by deprotonating lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues in the pdb file 4psw).The deprotonated lysine then acts as a nucleophile and attacks the carbonyl carbon of acetyl coenzyme Acetyl CoA (not shown in pdb file), forming a tetrahedral transition state with an oxyanion. The negative charge on the oxyanion then shift to down to reform the double bond between the oxygen and carbonyl carbon, breaking the scissle bond between the carbonyl carbon and the sulfur atom of acetyl CoA. The resulting product of this reaction is histone 4 with an acetyl-lysine at residue 12 and CoEnzyme A. | ||
| - | + | = Application = | |
| - | + | = References = | |
<references/> | <references/> | ||
Revision as of 22:21, 12 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||