We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Madeleine Wilson/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
===The Active Site=== | ===The Active Site=== | ||
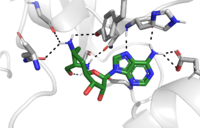
| - | The active site and binding pocket of KMT have several essential characteristics for the overall efficiency. First, the lysine of the histone enters the active site via the | + | The active site and binding pocket of KMT have several essential characteristics for the overall efficiency. First, the lysine of the histone enters the active site via the <scene name='81/811092/Tyrosine_channel_2/3'>Lysine access channel</scene> comprised of Tyr335 and Tyr337. Once in the active site, the alkyl part of the histone chain is stabilized by the <scene name='81/811092/Hydrophobic_binding_pocket/1'>hydrophobic binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. The Tyr335 and Tyr337 are also essential for stabilization of histone chain via hydrogen bonding. |
The <scene name='81/811092/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction. <ref name="Xiao" /> | The <scene name='81/811092/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction. <ref name="Xiao" /> | ||
[[Image:KMT_mechanism.png|200px|left|thumb|KMT Mechanism]] | [[Image:KMT_mechanism.png|200px|left|thumb|KMT Mechanism]] | ||
Revision as of 19:53, 24 April 2019
Histone Lysine Methyltransferase: Gene Activator
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ 5.0 5.1 5.2 5.3 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J Mol Biol. 1997 Jan 10;265(1):56-67. PMID:8995524 doi:http://dx.doi.org/10.1006/jmbi.1996.0711
- ↑ Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson