User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
=Histone Acetyltransferase HAT1/HAT2 Complex, ''Saccharomyces cerevisiae''= | =Histone Acetyltransferase HAT1/HAT2 Complex, ''Saccharomyces cerevisiae''= | ||
<StructureSection load='4PSW' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW' scene='81/811717/Overview/1'> | <StructureSection load='4PSW' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW' scene='81/811717/Overview/1'> | ||
| - | + | =Histones= | |
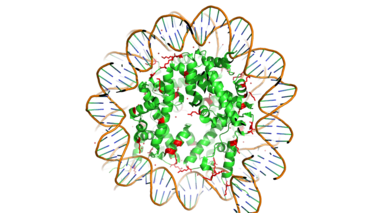
[https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins (H1A, H2A, H3, and H4) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] in which DNA directly interacts with and wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively-charged DNA is bound. <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref> It is those positively charged tails of the histone core that are often subject to post-translational modifications that play important roles in replication, transcription, heterochromatin maintenance, and DNA repair. | [https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins (H1A, H2A, H3, and H4) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] in which DNA directly interacts with and wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively-charged DNA is bound. <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref> It is those positively charged tails of the histone core that are often subject to post-translational modifications that play important roles in replication, transcription, heterochromatin maintenance, and DNA repair. | ||
[[Image:Histone_w_DNA.png|380 px|right|thumb|Figure 1. Histone Core w/ DNA Bound]] | [[Image:Histone_w_DNA.png|380 px|right|thumb|Figure 1. Histone Core w/ DNA Bound]] | ||
| - | + | =Histone Modification= | |
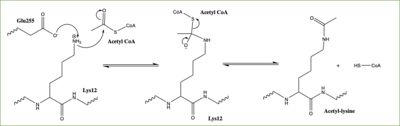
Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification that involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by the histone acetyltransferase (HAT) enzyme families. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. | Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification that involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by the histone acetyltransferase (HAT) enzyme families. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. | ||
Revision as of 01:24, 25 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||