User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
===Ligand Active Site=== | ===Ligand Active Site=== | ||
| - | [[Image:Hat1.gif|400 px|left|thumb|Figure | + | [[Image:Hat1.gif|400 px|left|thumb|Figure 2. Acetyl-CoA in the binding pocket of HAT1 (Chain A)]] |
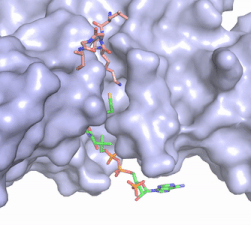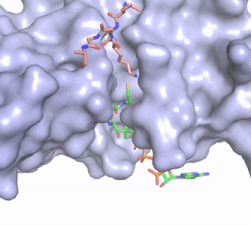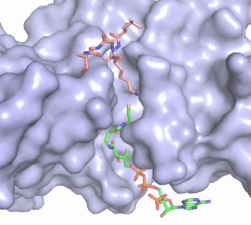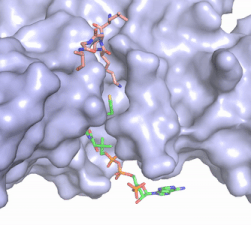
The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is buried in the protein, around 60% (Figure 1). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix α 7, C terminal end of strand β-14/loop β15-α9, and N-terminal half of helix α 9 <ref name=”Dutnall”>PMID:10384314</ref>. | The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is buried in the protein, around 60% (Figure 1). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix α 7, C terminal end of strand β-14/loop β15-α9, and N-terminal half of helix α 9 <ref name=”Dutnall”>PMID:10384314</ref>. | ||
| Line 30: | Line 30: | ||
Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. Previous studies found that a mutation at the active site glutamate residue greatly alters the catalytic ability of HAT1, proving it to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structural information from the crystallized structure of the HAT1/HAT2 complex regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactor: <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, H4Lys14, and Acetyl-CoA</scene>. | Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. Previous studies found that a mutation at the active site glutamate residue greatly alters the catalytic ability of HAT1, proving it to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structural information from the crystallized structure of the HAT1/HAT2 complex regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactor: <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, H4Lys14, and Acetyl-CoA</scene>. | ||
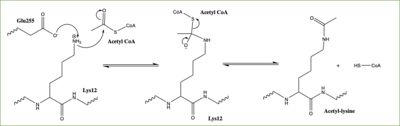
| - | [[Image:HAT1_Mechanism.jpg|400px|right|thumb|Figure | + | [[Image:HAT1_Mechanism.jpg|400px|right|thumb|Figure 3: Proposed HAT1 Arrow-Pushing Mechanism]] |
In this mechanism, the glutamate at residue 255 in the active site of the protein acts as a general base by deprotonating lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues in the pdb file 4psw).The deprotonated lysine then acts as a nucleophile and attacks the carbonyl carbon of Acetyl CoA (not shown in pdb file), forming a tetrahedral transition state containing an oxyanion. The negative charge on the oxyanion is then shift to down to reform the double bond between the oxygen and carbonyl carbon, breaking the scissle bond between the carbonyl carbon and the sulfur atom of acetyl CoA. The resulting product of this reaction is histone 4 with an acetyl-lysine at residue 12 and CoEnzyme A. | In this mechanism, the glutamate at residue 255 in the active site of the protein acts as a general base by deprotonating lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues in the pdb file 4psw).The deprotonated lysine then acts as a nucleophile and attacks the carbonyl carbon of Acetyl CoA (not shown in pdb file), forming a tetrahedral transition state containing an oxyanion. The negative charge on the oxyanion is then shift to down to reform the double bond between the oxygen and carbonyl carbon, breaking the scissle bond between the carbonyl carbon and the sulfur atom of acetyl CoA. The resulting product of this reaction is histone 4 with an acetyl-lysine at residue 12 and CoEnzyme A. | ||
Revision as of 18:24, 25 April 2019
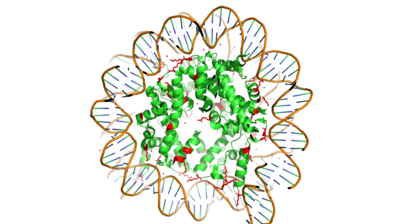
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||