This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
===Additional Features=== | ===Additional Features=== | ||
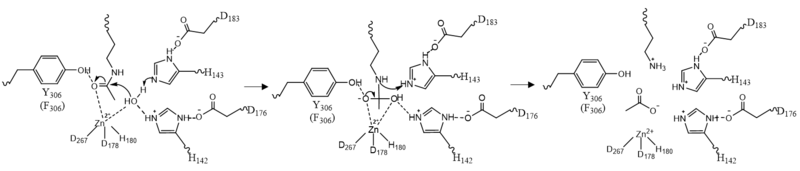
| - | There are two potassium ions bound in the HDAC8 structure. Potassium 1 is 7Å away from the active site while potassium 2 lies toward the exterior of the HDAC8.<ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref>. It is suggested that potassium 1 is of interest to the active site of HDAC8 because it is tethered to the enzyme by the main chain carbonyl oxygens of Asp178 and His180 which stabilizes the Zn<sup>2+</sup> in the active site. Furthermore, the potassium ion increases the positive charge in the active site and this could help stabilize the oxyanion hole that is formed in the transition state.<ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref> | + | There are two <scene name='81/811087/Potassium_ion/2'>potassium ions</scene> bound in the HDAC8 structure. Potassium 1 is 7Å away from the active site while potassium 2 lies toward the exterior of the HDAC8.<ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref>. It is suggested that potassium 1 is of interest to the active site of HDAC8 because it is tethered to the enzyme by the main chain carbonyl oxygens of Asp178 and His180 which stabilizes the Zn<sup>2+</sup> in the active site. Furthermore, the potassium ion increases the positive charge in the active site and this could help stabilize the oxyanion hole that is formed in the transition state.<ref name="Vannini, A., Volpari, C., Filocamo, G.">Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101</ref> |
<scene name='81/811087/Active_site_loop_1_s30-k36/11'>N-Terminus L1 loop </scene>(amino acid residues 30-36) makes up a significant part of the active site pocket. It is suggested that this loop has high flexibility that enables HDAC8 to more efficiently adjust binding to different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | <scene name='81/811087/Active_site_loop_1_s30-k36/11'>N-Terminus L1 loop </scene>(amino acid residues 30-36) makes up a significant part of the active site pocket. It is suggested that this loop has high flexibility that enables HDAC8 to more efficiently adjust binding to different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | ||
Revision as of 14:35, 26 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 3.4 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ 5.0 5.1 5.2 Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ 7.0 7.1 Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414