We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sean Callahan/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
LSD-1 will demethylate mono or di-methylated lysines. However it will not demethylate just any lysine. It will only demethylate lysine H3-K4. This factor can be regulated by the androgen receptor. The [http://proteopedia.org/wiki/index.php/Androgen_receptor#Function androgen receptor] is a protein that is involved in DNA transcription. When it interacts with LSD-1 it will no longer demethylate H3-K4, but will now demethylate H3-K9. This attribute allows LSD-1 to work on a wider range of residues<ref name="Stavropoulos">PMID: 16799558</ref>. | LSD-1 will demethylate mono or di-methylated lysines. However it will not demethylate just any lysine. It will only demethylate lysine H3-K4. This factor can be regulated by the androgen receptor. The [http://proteopedia.org/wiki/index.php/Androgen_receptor#Function androgen receptor] is a protein that is involved in DNA transcription. When it interacts with LSD-1 it will no longer demethylate H3-K4, but will now demethylate H3-K9. This attribute allows LSD-1 to work on a wider range of residues<ref name="Stavropoulos">PMID: 16799558</ref>. | ||
| - | + | ==Mechanism== | |
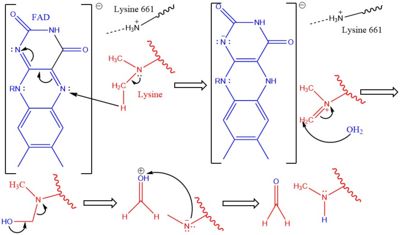
The demethylation of lysine via LSD1 occurs through oxidation via a hydride transfer. Shown in Figure 2 the first step involves the <scene name='81/811710/Fad_highlight/1'>Flavin Adenine Dinucleotide cofactor</scene> initiating a hydride transfer from the one of the two methyl groups bound to the nitrogen at the lysine tail, via a N5 on the flavin group. An imine cation is then formed at the end of the lysine tail to compensate for the loss of hydride. The FAD co-factor is also negatively charged and lysine residue 661 provides stability by drawing that charge away. In the next step the imine is hydrolyzed and transitions to an hemiaminal. This then breaks down to formaldehyde and the product lysine. | The demethylation of lysine via LSD1 occurs through oxidation via a hydride transfer. Shown in Figure 2 the first step involves the <scene name='81/811710/Fad_highlight/1'>Flavin Adenine Dinucleotide cofactor</scene> initiating a hydride transfer from the one of the two methyl groups bound to the nitrogen at the lysine tail, via a N5 on the flavin group. An imine cation is then formed at the end of the lysine tail to compensate for the loss of hydride. The FAD co-factor is also negatively charged and lysine residue 661 provides stability by drawing that charge away. In the next step the imine is hydrolyzed and transitions to an hemiaminal. This then breaks down to formaldehyde and the product lysine. | ||
[[Image:LSD1Mech2.jpg|400px|right|thumb|Figure 2]] | [[Image:LSD1Mech2.jpg|400px|right|thumb|Figure 2]] | ||
| - | + | ==Hydrophobic Pocket== | |
The hydrophobic pocket located in the active site cavity of LSD1, forms a catalytic chamber where the substrate lysine is oriented and positioned to interact with the | The hydrophobic pocket located in the active site cavity of LSD1, forms a catalytic chamber where the substrate lysine is oriented and positioned to interact with the | ||
FAD co-factor to initiate demethylation. The specific residues making up the pocket include valine-317, glycine-330, alanine-331, methionine 332, valine-333, phenylalanine-338, leucine-569, asparagine-660, lysine-661, tryptophan 695, serine 749, serine 760, and tyrosine-761. These residues in the <scene name='81/811712/Hydrophobic_pocket/4'>hydrophobic pocket</scene> shown in green surrounds the FAD in a way such that it exposes the catalytic nitrogen(N5) that is responsible for the two electron demethylation. The <scene name='81/811712/Fad_n5/3'>catalytic nitrogen</scene> depicted as a sphere is approximately 3.5Å away from the substrate lysine. Lysine 661 shown 4.98Å is responsible for anchoring the FAD in place to efficiently bind the substrate lysine. A structure complex of LSD1 with the a substrate lysine encased in has yet to be crystallized. Another three seperate pockets help bind the histone tail residues to the substrate lysine which is essential for identifying different modifications of the histone tail. | FAD co-factor to initiate demethylation. The specific residues making up the pocket include valine-317, glycine-330, alanine-331, methionine 332, valine-333, phenylalanine-338, leucine-569, asparagine-660, lysine-661, tryptophan 695, serine 749, serine 760, and tyrosine-761. These residues in the <scene name='81/811712/Hydrophobic_pocket/4'>hydrophobic pocket</scene> shown in green surrounds the FAD in a way such that it exposes the catalytic nitrogen(N5) that is responsible for the two electron demethylation. The <scene name='81/811712/Fad_n5/3'>catalytic nitrogen</scene> depicted as a sphere is approximately 3.5Å away from the substrate lysine. Lysine 661 shown 4.98Å is responsible for anchoring the FAD in place to efficiently bind the substrate lysine. A structure complex of LSD1 with the a substrate lysine encased in has yet to be crystallized. Another three seperate pockets help bind the histone tail residues to the substrate lysine which is essential for identifying different modifications of the histone tail. | ||
==Application== | ==Application== | ||
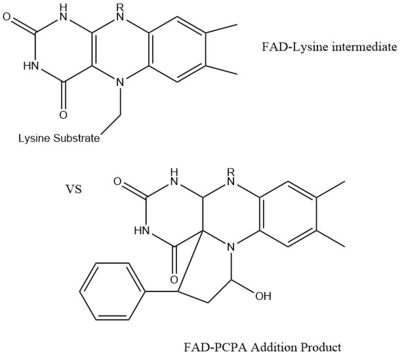
| - | LSD1 plays a | + | LSD1 plays a pivital role in many biological processes such as cell growth, epithelial-mesenchymal transition, stem cell biology, malignant transformation of cells, and cell differentiation. Malfunctioning of these processes can lead to life threatening diseases including but not limited to myeloid leukemia and acute lymphoblastic leukemia. Evidence has shown that these activities have ties to prostate and small cell lung cancer so studies have been done to find a reliable inhibitor for LSD1. Monamine oxidases utilize the FAD co-factor much like LSD1 and inhibitors of monamaine oxidase have proven to be successful in inhibiting the activity of LSD1. A patent for 5-cyano indole derivatives(compound similar in structure to monoamine oxidase inhibitors) has been proposed to be a potential inhibitor for LSD1. More specifically trans-2-Phenylcyclopropylamine has been shown the best representation of this inhibition. The formation of a product via an addition reaction through the flavin ring can restrict LSD1's activity with subsequent one‐electron oxidation and cyclopropyl ring opening. However due to the selective nature of 2-PCPA it does not present a strong enough affinity for LSD1's activity. Alternative structures using the foundation of 2-PCPA are continually being researched to find the best fit inhibitor for LSD1. An article published in Wiley Online Library discusses a number of potential LSD1 inhibitors based on monoamine oxidase inhibitors [https://onlinelibrary.wiley.com/doi/full/10.1002/med.21350]. Thus inhibition of LSD1 opens new pathways for possible treatments for cancers and disorders. |
| + | [[Image:FAD-PCPA.jpg|400px|right|thumb|Figure 2]] | ||
| + | |||
== References == | == References == | ||
<ref name=”Forneris">PMID: 16223729 </ref> | <ref name=”Forneris">PMID: 16223729 </ref> | ||
Revision as of 14:45, 26 April 2019
Lysine Specific Demethylase 1 (Homo sapiens)
| |||||||||||