We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
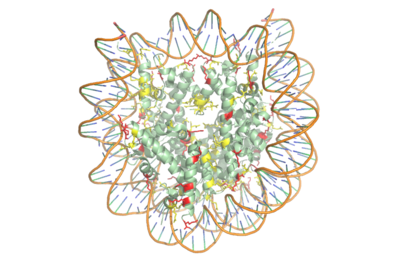
[https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins ([https://en.wikipedia.org/wiki/Histone_H1 H1], [https://en.wikipedia.org/wiki/Histone_H2A H2A], [https://en.wikipedia.org/wiki/Histone_H3 H3], and [https://en.wikipedia.org/wiki/Histone_H4 H4]) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] in which DNA directly interacts with and wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively-charged DNA is bound <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref> (Figure 1). After translation, the positively charged tails of the histone core that are often subject to modifications, such as acetylation or methylation. These modifications can regulate the processes of DNA repair, replication, transcription, and heterochromatin maintenance. | [https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins ([https://en.wikipedia.org/wiki/Histone_H1 H1], [https://en.wikipedia.org/wiki/Histone_H2A H2A], [https://en.wikipedia.org/wiki/Histone_H3 H3], and [https://en.wikipedia.org/wiki/Histone_H4 H4]) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] in which DNA directly interacts with and wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively-charged DNA is bound <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref> (Figure 1). After translation, the positively charged tails of the histone core that are often subject to modifications, such as acetylation or methylation. These modifications can regulate the processes of DNA repair, replication, transcription, and heterochromatin maintenance. | ||
| - | [[Image: | + | [[Image:Histone_NEW_w_DNA.png|400 px|right|thumb|Figure 1. Nucleosome consisting of the Histone core & DNA bound. Arginine residues shown in yellow. Lysine residues shown in red. PDB: 3kwq]] |
=Histone Modification= | =Histone Modification= | ||
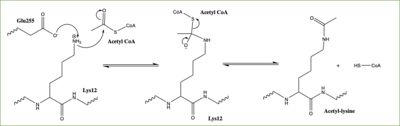
| - | Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is histone modification that involves the transfer of an acetyl group from Acetyl Coenzyme A (acetyl-CoA) to an ε-amino group of a lysine residue on a histone. This reaction is done by various histone acetyltransferase (HAT) enzymes. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV | + | Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is histone modification that involves the transfer of an acetyl group from Acetyl Coenzyme A (acetyl-CoA) to an ε-amino group of a lysine residue on a histone. This reaction is done by various histone acetyltransferase (HAT) enzymes. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. |
=HAT1 Background = | =HAT1 Background = | ||
| Line 37: | Line 37: | ||
= Inhibition = | = Inhibition = | ||
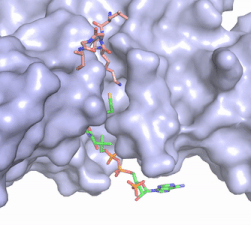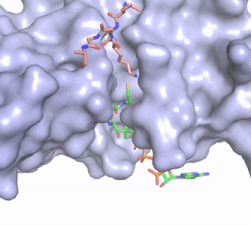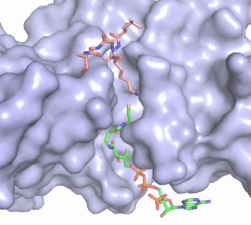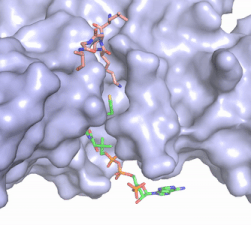
| - | Although HAT1 was the first histone acetyltransferase enzyme discovered, it is difficult to study and is one of the least understood HAT enzymes. While HAT1 has been linked to many disease states, there is no current known inhibitor of HAT1 that exists. Developing an enzyme inhibitor for HAT1 could allow for therapeutic targets in diseases in which HAT1 has been implicated as well as be used as a tool to better understand the specificity and mechanism in which HAT1 acts to modify histones, in particular histone 4 (H4). HAT1 inhibitors containing the first 20 residues of H4, including the target lysine for modification, and acetyl-CoA and found H4K12CoA to act as a competitive inhibitor to both the peptide substrate as well as acetyl-CoA, potentially laying the foundation for new discovery and better understanding of HAT1 <ref name="Ngo"/>. | + | Although HAT1 was the first histone acetyltransferase enzyme discovered, it is difficult to study and is one of the least understood HAT enzymes. While HAT1 has been linked to many disease states, there is no current known inhibitor of HAT1 that exists. Developing an enzyme inhibitor for HAT1 could allow for therapeutic targets in diseases in which HAT1 has been implicated as well as be used as a tool to better understand the specificity and mechanism in which HAT1 acts to modify histones, in particular histone 4 (H4). HAT1 inhibitors containing the first 20 residues of H4, including the target lysine for modification, and acetyl-CoA and found H4K12CoA to act as a competitive inhibitor to both the peptide substrate as well as acetyl-CoA, potentially laying the foundation for new discovery and better understanding of HAT1 <ref name="Ngo">PMID:30637990</ref>. |
= References = | = References = | ||
<references/> | <references/> | ||
Revision as of 17:37, 26 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||