This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
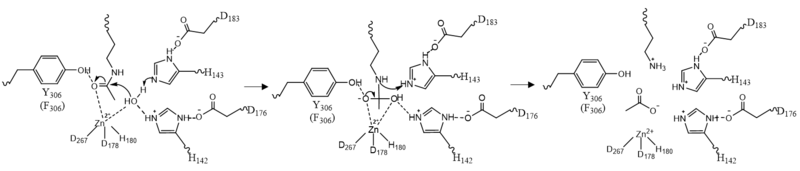
| - | Histone deacetylase 8 (HDAC8) is an enzyme that plays a role in controlling gene expression. Specifically, HDAC8 removes an acetyl group off of the ε-amino-Lys 382 of Histone 4's N-terminal core.<ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> [https://en.wikipedia.org/wiki/Histone Histones] consist of eight monomers to form an octomer complex. Each histone has a positive charge which allows interaction with negatively-charged DNA. This prevents transcription factors from accessing DNA, thus, decreasing gene expression. [https://en.wikipedia.org/wiki/Chromatin_remodeling Chromatin remodeling] by the addition or removal of an acetyl group on a histone is an example of [https://en.wikipedia.org/wiki/Epigenetics epigenetic regulation]. [https://en.wikipedia.org/wiki/Histone_acetyltransferase Histone Acetylase 1] (HAT1) catalyzes the addition of an acetyl group onto a histone. The lack of charge on the acetyl group weakens the interaction between DNA and histones which allows transcription factors to access | + | Histone deacetylase 8 (HDAC8) is an enzyme that plays a role in controlling gene expression. Specifically, HDAC8 removes an acetyl group off of the ε-amino-Lys 382 of Histone 4's N-terminal core.<ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> [https://en.wikipedia.org/wiki/Histone Histones] consist of eight monomers to form an octomer complex. Each histone has a positive charge which allows interaction with negatively-charged DNA. This prevents transcription factors from accessing DNA, thus, decreasing gene expression. [https://en.wikipedia.org/wiki/Chromatin_remodeling Chromatin remodeling] by the addition or removal of an acetyl group on a histone is an example of [https://en.wikipedia.org/wiki/Epigenetics epigenetic regulation]. [https://en.wikipedia.org/wiki/Histone_acetyltransferase Histone Acetylase 1] (HAT1) catalyzes the addition of an acetyl group onto a histone. The lack of charge on the acetyl group weakens the interaction between DNA and histones which allows transcription factors to access DNA to increase gene expression. HDAC8 reverses this reaction by catalyzing the removal of these acetyl groups from the Lys to reclaim the positive charge of the histone. This allows the histone to interact with the negative charge on the DNA. As a result, DNA binds more tightly to the histone protein, repressing transcription and gene expression. |
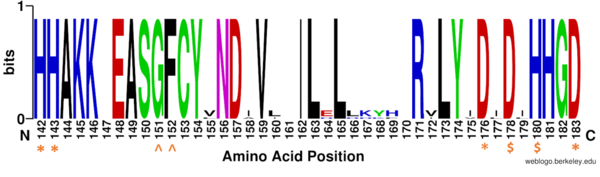
==HDAC Enzymes and Homology== | ==HDAC Enzymes and Homology== | ||
Revision as of 17:45, 26 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 3.4 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
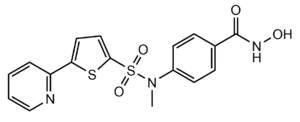
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ 5.0 5.1 5.2 Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ 7.0 7.1 Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414