We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Courtney Brown/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
===Histone Deacetylases (HDACs)=== | ===Histone Deacetylases (HDACs)=== | ||
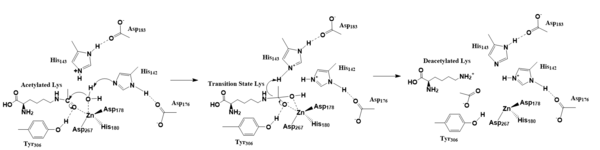
| - | HDACs are versatile enzymes that participate in regulating histone-DNA interactions, along with non-histone proteins, the cell cycle, cell differentiation and DNA-histone interactions<ref name="Vanninni" />. Histone acetylation is the reversal process, performed by [https://en.wikipedia.org/wiki/Histone_acetyltransferase HATs]. HDACs perform ε-amino-lysine deacetylation on acetylated lysines, the amount and residue number of which are unclear, within the histone<ref name="Seto">PMID:24691964</ref>. | + | HDACs are versatile enzymes that participate in regulating histone-DNA interactions, along with non-histone proteins, the cell cycle, cell differentiation and DNA-histone interactions<ref name="Vanninni" />. Histone acetylation is the reversal process of deacetylation by HDACs, performed by [https://en.wikipedia.org/wiki/Histone_acetyltransferase HATs]. HDACs perform ε-amino-lysine deacetylation on acetylated lysines, the amount and residue number of which are unclear, within the histone<ref name="Seto">PMID:24691964</ref>. |
There are different classes of HDACs based on phylogenetic analysis: Class I (HDACs 1-3 and 8), Class II (HDACs 4-7, 9 and 10), Class III (Sirtuin deacetylases), and Class IV (HDAC 11)<ref name="Vanninni">PMID:17721440</ref>. These different classes share residues that are largely conserved throughout (Figure 1). These conserved residues are either in the active site or involved in substrate binding, as HDACs 1-11 are metalloenzymes with similar reaction mechanisms. They all utilize a zinc ion in their deacetylation<ref name="Vanninni" />. [[Image:Screen_Shot_2019-04-08_at_12.37.29_AM.png|600 px|center|thumb|Figure 1. Conserved Residues Across HDAC Families. Red represents His142 and His143, purple represents Asp178, blue represents His180, pink represents Asp101, yellow represents Tyr306 and green represents Asp267.]] | There are different classes of HDACs based on phylogenetic analysis: Class I (HDACs 1-3 and 8), Class II (HDACs 4-7, 9 and 10), Class III (Sirtuin deacetylases), and Class IV (HDAC 11)<ref name="Vanninni">PMID:17721440</ref>. These different classes share residues that are largely conserved throughout (Figure 1). These conserved residues are either in the active site or involved in substrate binding, as HDACs 1-11 are metalloenzymes with similar reaction mechanisms. They all utilize a zinc ion in their deacetylation<ref name="Vanninni" />. [[Image:Screen_Shot_2019-04-08_at_12.37.29_AM.png|600 px|center|thumb|Figure 1. Conserved Residues Across HDAC Families. Red represents His142 and His143, purple represents Asp178, blue represents His180, pink represents Asp101, yellow represents Tyr306 and green represents Asp267.]] | ||
One type of HDAC, [https://en.wikipedia.org/wiki/HDAC8 Histone Deacetylase 8], is an enzyme found in ''[https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]''. HDAC8 decreases gene expression by increasing DNA-histone interactions, condensing the DNA tighter, into the transcriptionally silent heterochromatin<ref name="Vanninni" />. | One type of HDAC, [https://en.wikipedia.org/wiki/HDAC8 Histone Deacetylase 8], is an enzyme found in ''[https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]''. HDAC8 decreases gene expression by increasing DNA-histone interactions, condensing the DNA tighter, into the transcriptionally silent heterochromatin<ref name="Vanninni" />. | ||
Revision as of 23:40, 26 April 2019
The Human Histone Deacetylase, HDAC8
| |||||||||||
References
- ↑ 1.0 1.1 Histones | Learn Science at Scitable https://www.nature.com/scitable/definition/histone-histones-57
- ↑ What is chromatin, heterochromatin and euchromatin? MBInfo https://www.mechanobio.info/genome-regulation/what-is-chromatin-heterochromatin-and-euchromatin
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007 Sep;8(9):879-84. Epub 2007 Aug 10. PMID:17721440
- ↑ Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi:, 10.1101/cshperspect.a018713. PMID:24691964 doi:http://dx.doi.org/10.1101/cshperspect.a018713
- ↑ Chen K, Zhang X, Wu YD, Wiest O. Inhibition and mechanism of HDAC8 revisited. J Am Chem Soc. 2014 Aug 20;136(33):11636-43. doi: 10.1021/ja501548p. Epub 2014, Aug 7. PMID:25060069 doi:http://dx.doi.org/10.1021/ja501548p
- ↑ Tabackman AA, Frankson R, Marsan ES, Perry K, Cole KE. Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket. J Struct Biol. 2016 Sep;195(3):373-8. doi: 10.1016/j.jsb.2016.06.023. Epub 2016, Jun 29. PMID:27374062 doi:http://dx.doi.org/10.1016/j.jsb.2016.06.023
- ↑ 7.0 7.1 Marks PA. Histone deacetylase inhibitors: a chemical genetics approach to understanding cellular functions. Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):717-25. doi:, 10.1016/j.bbagrm.2010.05.008. Epub 2010 Jun 8. PMID:20594930 doi:http://dx.doi.org/10.1016/j.bbagrm.2010.05.008
- ↑ Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004 Oct 19;101(42):15064-9. Epub 2004 Oct 11. PMID:15477595
Student Contributors
- Courtney Brown
- Cassandra Marsh
- Carolyn Hurdle