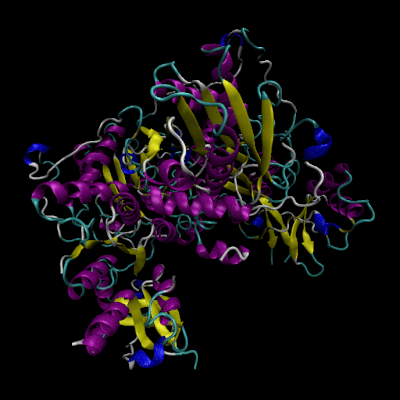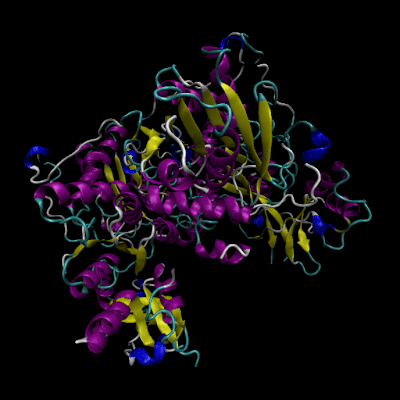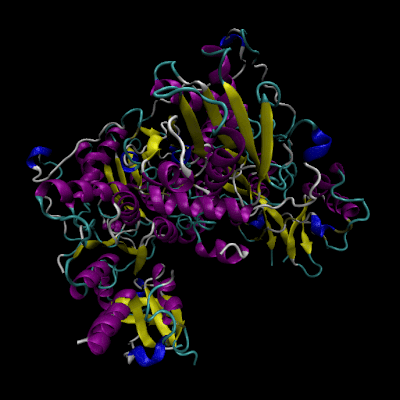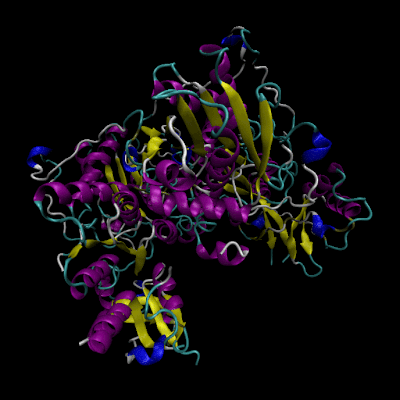
Cystathionine β-synthase (CBS; EC 4.2.1.22) is a unique pyridoxal 5’-phosphate (PLP) enzyme which plays a crucial role in the transsulfuration metabolic pathway of sulfur-containing amino acid, L-homocysteine.
PLP enzymes are divided into four families on the basis of similarities in three dimensional structure, sequence, secondary structure, and hydrophobicity profiles. CBS is a member of the β family or Fold type II, which also contains O-acetylserine sulfhydrylase (OASS) and O-acetyl-L-serine(thiol)lyase (OASTL)[1].
Structure of CBS
The human form of CBS is a homotetramer consisting of 63-kDa subunits each of 551 amino acids in length. Two cofactors, pyridoxal 5’-phosphate (PLP) and heme, and two substrates, L-homocysteine and L-serine, are bound to every monomer and the function is further allosterically regulated by S-adenosyl-L-methionine (AdoMet).
The monomer has a complex domain structure as it is composed of three functional domains, two of which are regulatory and one is catalytic. The middle domain contains the catalytic core responsible for the pyridoxal 5’-phosphate-catalyzed reaction and is flanked by an N-terminal heme-binding domain (70 amino acids), which probably regulates the enzyme in response to redox conditions, and a C-terminal Bateman module consisting of two CBS domains (CBS1, CBS2), protein folding motifs also found in inosine 5’-monophosphate dehydrogenase, chloride channels and several other proteins in various organisms.[2] The C-terminal domain also contains a negative regulatory region that is responsible for allosteric activation of the enzyme by AdoMet.
Truncation of N-terminal domain produces an enzyme that is still active albeit less so (19%) than the wild type. Truncation of C-terminal domain activates the enzyme by ~2-fold over full-length wild type. In addition, deletion of the Bateman module leads to a change in oligomerization from a tetrameric to a dimeric form of the enzyme.[3][4]
The CBS tetramer of the full-length enzyme has a strong tendency to aggregate, which makes physical studies very difficult. Therefore recombinant human CBS comprising the amino acid residues 1±413 (C-terminal domain missing), which does not exhibit the aggregating properties, is described.
The monomer is composed of 11 α-helices, seven short 310 helices and two β-sheets consisting of four (in the N-terminal domain) and six strands (in the C-terminal domain), respectively. The additional β-strand 1 interacts with strand 2 of the C-terminal sheet of the other monomer in the dimer in a parallel manner. The heme binding motif lacks any secondary structure with the exception of a short 310 helix.
The dimer interface is mainly hydrophobic in character and is composed of the side chains of the residues Ile76, Leu77, Ile80, Thr87, Val90, Ile92, Ile152, Leu156, Val160, Val180, Ala183, Leu184, Ile339, Ala340, Leu344, Leu345, Val378, Met382 and Leu386.
The central part of the dimer interface is formed by the residues Phe111 and Phe112 close to the 2-fold dimer axis, thus Phe112 of monomer A interacts with Phe112 of monomer B and vice versa. Polar interactions also contribute to the dimer interactions. On the other hand, the guanidium group of Arg379 is completely buried within the core of the dimer interface and rather than involved in any polar interactions between monomers forms hydrogen bonds to Gly115 and Asn380 of the same monomer.[5]
The active site
The coenzyme PLP is deeply buried in a cleft between the N- and C-terminal domains, and the active site is accessible only via a narrow channel. The cofactor is linked to the ε-amino group of Lys119 via a Schiff base linkage forming the so-called 'internal aldimine'. The nitrogen of the pyridine ring forms a hydrogen bond to the Oγ of Ser349. Another hydrogen bond is formed between the 3’ hydroxyl group of PLP and the Nδ2 of Asn149. This residue is coplanar with the pyridine ring and thus allows the expected ring tilt upon transaldimination.
The phosphate binding loop is located between β-strand 8 and α-helix 8 and is composed of the residues Gly256, Thr257, Gly258, Gly259 and Thr260. These residues form an extended hydrogen bonding network with the phosphate moiety of PLP, thus anchoring the cofactor to the protein matrix. In addition, the positive pole of the helix dipole from α-helix 8 compensates for the negative charge of the phosphate group.
Residues Tyr223 and Gly307 are probably the key residues for substrate specificity, as they are spatially adjacent to the substrate binding site. After binding substrate the catalytic core undergoes a conformational change.[6]
The heme binding site
Heme molecules are located at distal ends of the dimers with the orientation of their ring planes normal to the protein surface. The heme is bound in a hydrophobic pocket formed by residues 50±67, α-helices 6 and 8 and a loop preceding β-strand 10. The sulfhydryl group of Cys52 and the Nε2 atom of His65 axially coordinate the iron in the heme. The sulfur atom of Cys52 is deprotonated and forms additional polar interactions with the side chain of Arg266 and the main chain nitrogen of Trp54. Nε2 of His65 is solvent accessible and lacking any hydrogen bonding partner from protein residues.
The heme carboxylate groups are involved in ionic interactions with Arg51 and Arg224 and are partially solvent accessible.
Since the iron ion is ligated from both sides by protein residues this makes an enzymatic role of the heme unlikely. There is no covalent attachment of the heme to the protein which means it can be reversibly released under reducing conditions in the presence of carbon monoxide (CO). Under oxidizing conditions, the heme cannot be released, probably because CO does not bind to heme in its ferric state.
It is likely that CO displaces one of the axial heme ligands, followed by a local unfolding of the N-terminal residues leading to a release of the prosthetic group. The displaced ligand is probably the cysteine, because the absorption spectrum of CBS treated with CO is similar to the spectra of other CO±heme±imidazole protein complexes.[7]
Oxidoreductase active site motif
The loop between α-helix 8 and β-strand 9 harbors a motif similar to the active site of disulfide oxidoreductases. In CBS this motif consists of the sequence CPGC (residues 272±275) and forms a β-turn. The two cysteines are oxidized and form a disulfide bridge. The disulfide bridge is in a right-handed hook conformation and is located on the surface of the protein and hence is solvent accessible. The same two cysteines, however, are not solvent accessible in the full-length enzyme.[8]
The regulatory domain
Full-length CBS contains a C-terminal regulatory domain of ~140 residues, including two so-called 'CBS domains' (CBS1 of 53 residues and CBS2 of 57 residues). The C-terminal domain of CBS contains an autoinhibitory region that gets displaced from the active site upon binding of the allosteric activator AdoMet.
The fact that truncated CBS forms dimers rather than tetramers or higher order oligomers suggests that the regulatory domain is involved in tetramer formation.
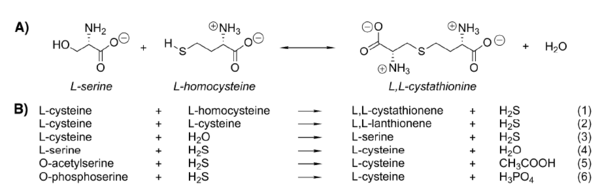
The Reaction Catalyzed by CBS
Cystathionine β-synthase catalyzes β-elimination and β-replacement reactions. In a typical situation it condensates L-serine and L-homocysteine to give cystathionine but there are also other possible substrates for this enzyme leading to different products.
The reaction for synthesis of cystathionine starts with displacement of the internal aldimine between the enzyme active site lysine and pyridoxal 5’-phosphate (E-PLP) by the incoming L-serine. The serine external aldimine adduct (E-PLP-L-ser) forms an aminoacrylate intermediate (E-PLP-aa) that reacts with the incoming second substrate, such as L-homocysteine, to form the (L,L)-cystathionine external aldimine, which is then displaced by the active site lysine to regenerate the active enzyme.[9]
The type of reaction mechanisms used by the CBS is known as a double displacement or ping-pong mechanism. The rate determining step in the reaction is hydrolysis of the external aldimine of cystathionine.[10]

Mutations in CBS gene
There are more than 100 reported mutations in CBS gene from patients with homocystinuria most of which are missense. Among most frequent mutations is I278T (c.833T>C) in C-terminal domain found in about 25 % of all homocystinuric alleles, and G307S (c.919G>A) located in active site.
Mutations in active site
The active site is accessible only via a narrow channel. Four of the six known point mutations in the active site involve glycine residues: G148R, G305R, G307S and G259S. The residue G259 separates the active site from the heme-binding pocket. The residue G307 lines the entry to the active site cleft and the orientation of G307 do not allow accommodating of the side chain of a serine residue which causes incorporation of the side chain and conformation change in the loop. As the second substrate homocysteine probably binds here the mutation could inhibit binding of homocysteine.
Mutations in the heme-binding site
Mutations in this part of the enzyme reduces the ability to bind heme and affects proper folding of CBS.
Mutations in the dimer interface
Numerous CBS mutations (e.g. A114V and G116R) are located at interface of the two monomers and destabilise monomer-monomer interactions and their communication.
Other mutations
Residues in regulatory domain (414-551) bind the allosteric activator S-adenosyl-L-methionine and are responsible for the tetramerization. Mutation R336/H belong to this group of mutations – the side chain of this arginine is packed against the protein surface and the guanidium groups forms a salt bridge to the carboxyl group of D388, residue that does not contribute to any interaction between the two monomers.
The most common mutation, I278T, is located in β-sheet of the C-terminal domain.
Disease
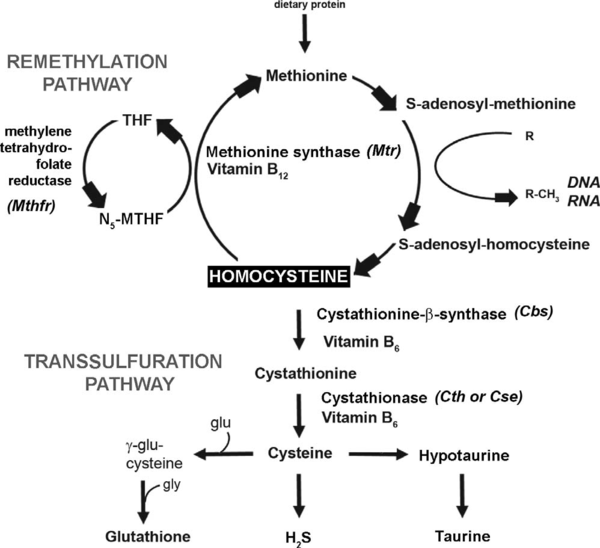
The Metabolic Pathway of Homocysteine
The metabolism of L-methionine comprises two intersecting pathways: the synthesis pathway via the S-adenosyl methionine cycle (SAM cycle) and the transsulfuration pathway leading to glutathione synthesis. At the intersection of these sits homocysteine (Hcy), a non-proteinogenic amino acid whose elevated plasma levels (eHcy) has been observed in several medical conditions indicating the toxic effect of this chemical substance and need for rigorous control of its amount.
The Hcy concentrations are maintained dynamically by either a transsulfuration or remethylation pathway provided by different enzymes.
The remethylation reaction is carried out by two enzymes. Methionine synthase, with vitamin B12 as a cofactor, remethylates Hcy back to methionine. Methylene tetrahydrofolate (MTHF) derived from tetrahydrofolate (THF) by methylenetetrahydrofolate reductase (MTHFR) is used as a donor of the methyl group so folate is also required for this reaction. The remethylation pathway is favored under conditions where methionine levels are low, resulting in conservation of this metabolite.
When methionine levels are high, transsulfuration dominates. The transsulfuration pathway catalyzes the conversion of homocysteine to cysteine, and is the only de novo pathway for cysteine production in mammals. In the first step, homocysteine is condensed with serine to form cystathionine, the reaction is catalyzed by cystathionine β-synthase (CBS), and is dependent on vitamin B6 as an enzymatic cofactor. In the second step, cystathionine is cleaved by cystathionase (CTH), also known as cystathionine γ-lyase (CSE), to form cysteine and α-ketobutyrate. Cysteine is used in synthesis of downstream products such as glutathione (GSH), taurine, and hydrogen sulfide (H2S).
Hcy is a precursor of S-adenosyl-L-methionine (AdoMet), a methyl group donor in a large number of biochemical reactions, and a metabolite of S-adenosyl-L-homocysteine (AdoHcy). The ratio of AdoMet to AdoHcy is defined as the methylation potential (MP). The two pathways are coordinated by AdoHcy, which acts as an allosteric inhibitor of the MTHFR reaction and as an activator of CBS.
Among the pathological states that have been mentioned in relation with eHcy are cardiovascular disorders, atherosclerosis, myocardial infarction, stroke, minimal cognitive impairment, dementia, Parkinson’s disease, Alzheimer’s disease, multiple sclerosis, epilepsy, and eclampsia.[11] All these observations indicates that Hcy, and especially eHcy, exerts direct toxic effects on both the vascular and nervous systems.
Mutations in the gene encoding CBS resulting in abnormalities of its function with all the consequences are usually referred as of homocystinuria. The mutations can alter either mRNA or enzyme stability, activity, binding of PLP and heme, or impair allosteric regulation.[12] To date there have been over 100 mutations described in this gene.[13]
Deficiency in the activity of the enzyme caused by insufficient supplementation of enzymatic cofactores leads to accumulation of L-homocysteine, hyperhomocysteinemia, with similar but moderate indications like homocystinuria.
Involvement in such a wide number of apparently unrelated diseases may be caused by affecting a very basic biological process central to a variety of diseases. There are two major hypotheses about how this could be accomplished.
The first one has centered on the relationship between homocysteine and oxidative stress. Homocysteine itself has been shown to cause increased oxidative stress on cells, both through direct effects (e.g., the production of hydrogen peroxide by oxidation of homocysteine to homocystine) and indirect effects (e.g., reduction of glutathione peroxidase). In addition, it is estimated that as much as 50% of the cellular antioxidant glutathione is produced from homocysteine by conversion through the transsulfuration pathway.
A second popular hypothesis suggests that eHcy affects the control of biologically important methylation reactions by causing a build-up of S-adenosyl-L-homocysteine (AdoHcy) which is a competitive inhibitor of S-adenosyl-L-methionine (AdoMet) binding for methyltransferase enzymes. As methyltransferases are involved in a variety of important biological processes, inhibition of this class of enzymes could have extremely diverse effects on the organism.[14]
Molecular dynamic simulation
For this project we also include animated gif files of our very own molecular dynamic simulation of the solvated and ionized CBS. Movement of the enzyme has been computed by CHARMM in MetaCentrum VO. Input has been prepared in VMD from 4coo PDB file.
Animation should be visible upon clicking on the images.