Histidine triad nucleotide-binding protein
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Structural highlights == | == Structural highlights == | ||
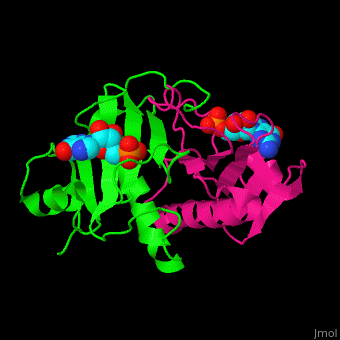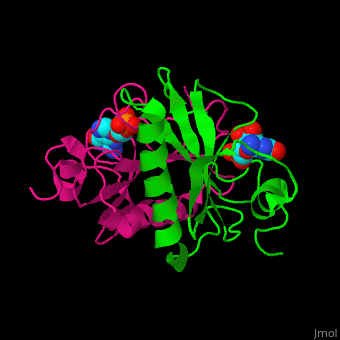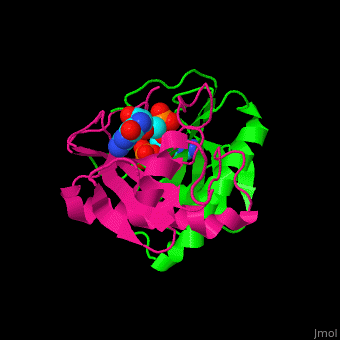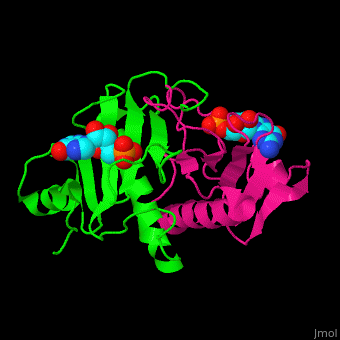
The <scene name='59/593304/Cv/5'>HINT nucleotide-binding cleft is at the top of the β sheet</scene><ref>PMID:9164465</ref>. Water molecules shown as red spheres. | The <scene name='59/593304/Cv/5'>HINT nucleotide-binding cleft is at the top of the β sheet</scene><ref>PMID:9164465</ref>. Water molecules shown as red spheres. | ||
| + | |||
| + | ==3D structures of histidine triad nucleotide-binding protein== | ||
| + | [[Histidine triad nucleotide-binding protein 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| Line 20: | Line 24: | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | *HINT1 | + | *HINT1 or adenosine 5’-monophosphoramidase |
| - | **[[1kpa]], [[1kpb]], [[1kpc]] – hHINT – human<br /> | + | **[[1kpa]], [[1kpb]], [[1kpc]], [[6g9z]], [[5o8i]], [[4zkv]] – hHINT – human<br /> |
| + | **[[5waa]], [[5ipb]] – hHINT (mutant) <br /> | ||
**[[6rhn]] – rHINT - rabbit<br /> | **[[6rhn]] – rHINT - rabbit<br /> | ||
**[[3o1z]] – rHINT (mutant) <br /> | **[[3o1z]] – rHINT (mutant) <br /> | ||
| Line 28: | Line 33: | ||
*HINT1 complexes | *HINT1 complexes | ||
| - | **[[3rhn]] – rHINT + GMP <br /> | ||
| - | **[[4rhn]], [[3qgz]] – rHINT + adenosine<br /> | ||
| - | **[[5rhn]] – rHINT + 8-Br-AMP <br /> | ||
**[[1av5]], [[1kpe]] – hHINT + adenosine derivative<br /> | **[[1av5]], [[1kpe]] – hHINT + adenosine derivative<br /> | ||
| - | **[[1kpf]], [[3tw2]] – hHINT + | + | **[[1kpf]], [[3tw2]], [[6b42]], [[5klz]], [[5km0]], [[5km1]], [[5km2]], [[5km3]], [[5ed3]] – hHINT + nucleotide <br /> |
| - | **[[ | + | **[[5ed6]] – hHINT (mutant) + nucleotide <br /> |
| - | **[[ | + | **[[5km4]], [[4zkl]] – hHINT + nucleotide derivative<br /> |
| + | **[[5ipd]], [[5ipe]] – hHINT + phosphoramidate derivative<br /> | ||
| + | **[[5kly]], [[5km6]], [[5kma]], [[5ipc]] – hHINT (mutant) + phosphoramidate derivative<br /> | ||
**[[4eqe]] – hHINT + Lys-AMS <br /> | **[[4eqe]] – hHINT + Lys-AMS <br /> | ||
**[[4eqg]] – hHINT + Ala-AMS <br /> | **[[4eqg]] – hHINT + Ala-AMS <br /> | ||
| + | **[[5kmc]] – hHINT + adenosine-phosphoramidate <br /> | ||
| + | **[[5wa8]], [[5wa9]] – hHINT (mutant) + Ala-phosphoramidate <br /> | ||
| + | **[[5kmb]] – hHINT (mutant) + Trp-phosphoramidate <br /> | ||
**[[4eqh]] – hHINT + Trp-AMS <br /> | **[[4eqh]] – hHINT + Trp-AMS <br /> | ||
| + | **[[5i2e]], [[5i2f]] – hHINT + sulfamate inhibitor<br /> | ||
| + | **[[5emt]] – hHINT + Cu inhibitor <br /> | ||
| + | **[[3rhn]] – rHINT + GMP <br /> | ||
| + | **[[4rhn]], [[3qgz]] – rHINT + adenosine<br /> | ||
| + | **[[1rzy]], [[5rhn]] – rHINT + adenosine derivative<br /> | ||
| + | **[[3o1c]], [[3o1x]] – rHINT (mutant) + adenosine <br /> | ||
*HINT2 | *HINT2 | ||
**[[4inc]], [[4njx]], [[4njy]], [[4njz]], [[4nk0]] – hHINT <br /> | **[[4inc]], [[4njx]], [[4njy]], [[4njz]], [[4nk0]] – hHINT <br /> | ||
| + | **[[5km9]] – hHINT + adenosine<br /> | ||
| + | **[[6km8]] – hHINT + cidofovir<br /> | ||
| + | **[[6km5]] – hHINT + triciribine derivative<br /> | ||
**[[4ini]] – hHINT + AMP <br /> | **[[4ini]] – hHINT + AMP <br /> | ||
Revision as of 09:59, 25 July 2019
| |||||||||||
3D structures of histidine triad nucleotide-binding protein
Updated on 25-July-2019
References
- ↑ Linde CI, Feng B, Wang JB, Golovina VA. Histidine triad nucleotide-binding protein 1 (HINT1) regulates Ca(2+) signaling in mouse fibroblasts and neuronal cells via store-operated Ca(2+) entry pathway. Am J Physiol Cell Physiol. 2013 Jun 1;304(11):C1098-104. doi:, 10.1152/ajpcell.00073.2013. Epub 2013 Apr 10. PMID:23576580 doi:http://dx.doi.org/10.1152/ajpcell.00073.2013
- ↑ Wistuba II, Ashfaq R, Maitra A, Alvarez H, Riquelme E, Gazdar AF. Fragile histidine triad gene abnormalities in the pathogenesis of gallbladder carcinoma. Am J Pathol. 2002 Jun;160(6):2073-9. PMID:12057912 doi:http://dx.doi.org/10.1016/S0002-9440(10)61157-1
- ↑ Zochbauer-Muller S, Wistuba II, Minna JD, Gazdar AF. Fragile histidine triad (FHIT) gene abnormalities in lung cancer. Clin Lung Cancer. 2000 Nov;2(2):141-5. PMID:14731325
- ↑ Wierzbicki PM, Adrych K, Kartanowicz D, Wypych J, Stanislawowski M, Zwolinska-Wcislo M, Celinski K, Skrodzka D, Godlewski J, Korybalski B, Smoczynski M, Kmiec Z. Overexpression of the fragile histidine triad (FHIT) gene in inflammatory bowel disease. J Physiol Pharmacol. 2009 Oct;60 Suppl 4:57-62. PMID:20083852
- ↑ Wierzbicki PM, Adrych K, Kartanowicz D, Dobrowolski S, Stanislawowski M, Chybicki J, Godlewski J, Korybalski B, Smoczynski M, Kmiec Z. Fragile histidine triad (FHIT) gene is overexpressed in colorectal cancer. J Physiol Pharmacol. 2009 Oct;60 Suppl 4:63-70. PMID:20083853
- ↑ Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins. Nat Struct Biol. 1997 Mar;4(3):231-8. PMID:9164465