User:Luis Andres Casavilca Ramirez/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
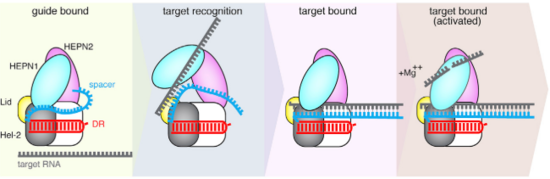
| - | The crRNA is practically inaccessible for hybridization with target RNA unless a major conformational shift allows access to the central channel. An opening between the domains HEPN1 and Helical-2 has been suggested, | + | The Cas13b-crRNA complex is practically inaccessible for hybridization with target RNA unless a major conformational shift allows access to the central channel. An opening between the domains HEPN1 and Helical-2 has been suggested, since it would provide a sterically permissible route with charged amino acids that would direct the RNA to the Cas13b central cavity. Mutation in a residue on a Lid domain β-hairpin (D397) at the interface between HEPN1 and Helical2 decreases Cas13 knockdown activity, suggesting an additional role of this domain in directing the target RNA into the central channel. |
| - | [[Image:Selection_039.png|550px|right|thumb| Fig.2 Proposed of crRNA targeting by Cas13b]] | + | [[Image:Selection_039.png|550px|right|thumb| Fig.2 Proposed mechanism of crRNA targeting by Cas13b]] |
The 3’end of target RNA is less tightly bound within the central channel and more tolerant of mismatches that the 5’end, thus being proposed as being initially recognized by the complex. In this suggested model the protein-crRNA complex initially probes the 3’RNA end of target RNA, allowing the opening of HEPN1 and Helical-2 domains and access to the central channel after complementarity is found. Then, the rest of the target RNA is hybridized. (Fig.2) | The 3’end of target RNA is less tightly bound within the central channel and more tolerant of mismatches that the 5’end, thus being proposed as being initially recognized by the complex. In this suggested model the protein-crRNA complex initially probes the 3’RNA end of target RNA, allowing the opening of HEPN1 and Helical-2 domains and access to the central channel after complementarity is found. Then, the rest of the target RNA is hybridized. (Fig.2) | ||
| Line 42: | Line 42: | ||
| - | == | + | == Cas13b engineering == |
| + | |||
| + | The aforementioned preference for target RNA cleavage at dinucleotide sites is somewhat conserved among Cas 13 enzymes. A beta-hairpin loop located near the active site at HEPN2 domain is conserved among Cas13 from different species. Though the identities of its residues are variable, it is always located between highly conserved amino-acids (2) In PbuCas13b, mutants with altered residues at this loop that retained cleavage activity showed different cleavage preferences, and some of them even had increased preference for UU dinucleotides. SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing), an mRNA detection technique that takes advantage of the promiscuous cleavage activity of Cas13s is limited by the nucleobases identities of some of transcripts.(8) Therefore, Cas13b engineering at this conserved loop could potentially increase the repertoire of substrates for these type of techniques. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | 1. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80- ). 2016;353(6299). | ||
| - | 2. Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, et al. High-Resolution Structure of Cas13b and Biochemical Characterization of RNA Targeting and Cleavage. Cell Rep. 2019 Mar 26;26(13):3741-3751.e5. | ||
| - | 3. Wolter F, Puchta H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94(5):767–75. | ||
| - | 4. Brennicke A. RNA editing [Internet]. Vol. 23, FEMS Microbiology Reviews. 1999. 297–316 p. Available from: http://doi.wiley.com/10.1016/S0168-6445(99)00009-1 | ||
| - | 5. Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell [Internet]. 2017;65(4):618-630.e7. Available from: http://dx.doi.org/10.1016/j.molcel.2016.12.023 | ||
| - | 6. East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature [Internet]. 2016;538(7624):270–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27669025%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5576363 | ||
| - | 7. Yang W. Nucleases: diversity of structure, function and mechanism. Vol. 44, Quarterly Reviews of Biophysics. 2011. 1–93 p. | ||
Revision as of 05:26, 17 June 2019
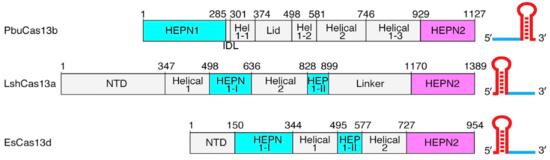
==Cas13b==
| |||||||||||