User:Luis Andres Casavilca Ramirez/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <scene name='81/817991/Pbucas13b_central_channel/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_domain/2'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat2-33-36/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_u20/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_pocket/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat/3'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_a/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat/1'>Text To Be Displayed</scene><scene name='81/817991/Crrna/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_active_site/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_beta_hairpin/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn2_active_site/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn_site/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn2/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn1/1'>Text To Be Displayed</scene><scene name='81/817991/Helical_1_and_helical_2/4'>Text To Be Displayed</scene><scene name='81/817991/Helical_1_and_helical_2/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn1_and_hepn2/1'>Text To Be Displayed</scene><scene name='81/817991/Pbucas13b_domains/2'>Text To Be Displayed</scene>==Cas13b== | + | <scene name='81/817991/Hepn_siteb/1'>Text To Be Displayed</scene><scene name='81/817991/Pbucas13b_central_channel/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_domain/2'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat2-33-36/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_u20/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_pocket/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat/3'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat_a/1'>Text To Be Displayed</scene><scene name='81/817991/Rna_direct_repeat/1'>Text To Be Displayed</scene><scene name='81/817991/Crrna/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_active_site/1'>Text To Be Displayed</scene><scene name='81/817991/Lid_beta_hairpin/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn2_active_site/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn_site/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn2/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn1/1'>Text To Be Displayed</scene><scene name='81/817991/Helical_1_and_helical_2/4'>Text To Be Displayed</scene><scene name='81/817991/Helical_1_and_helical_2/1'>Text To Be Displayed</scene><scene name='81/817991/Hepn1_and_hepn2/1'>Text To Be Displayed</scene><scene name='81/817991/Pbucas13b_domains/2'>Text To Be Displayed</scene>==Cas13b== |
<StructureSection load='6dtd' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='6dtd' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| Line 16: | Line 16: | ||
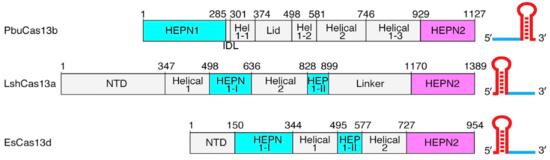
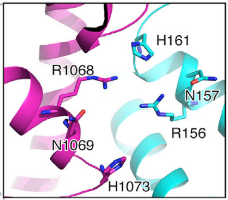
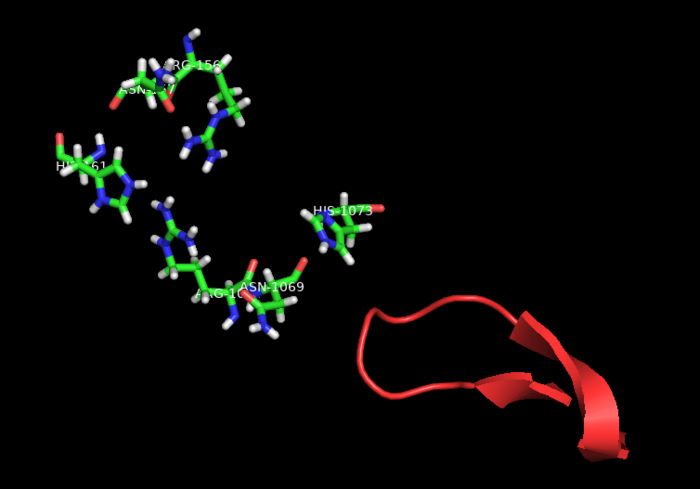
| - | HEPNs (Higher Eukaryotes and Prokaryotes Nucleotide-binding domain) are alpha helical domains, many of which are present in RNA maturation systems and related biological conflicts and most of which contain a catalytic Rx4-6H active site. Cas 13b <scene name='81/817991/Hepn1/1'>HEPN1</scene> domain is composed of 12 linearly connected alpha helices flexible loop between them, whereas <scene name='81/817991/Hepn2/1'>HEPN2</scene> contains nine alpha helices, several short beta strands and beta-hairpin with positively charged residues at the tip. In accordance with the conserved active site from HEPNs, a Rx4-6H site is present, where active residues locate within both HEPN1 (<scene name='81/817991/Hepn1_active_site/1'>R156, N157, H161</scene>) and HEPN2 (<scene name='81/817991/Hepn2_active_site/1'>R1068, N1069, H1073</scene>) domains. One of the two histidines is thought to act as a base, inducing the ribose 2’OH to attack the phosphodiester linkage. The conserved arginine could stabilize the negative oxygens of the transition state, as a similar residue (Lysine) has been shown to carry out such function in RNase A. Alternatively, it could interact with the RNA backbone. The other polar residues, located in between the catalytic histidine and the conserved arginine, are thought to further augment the active site. (2,4) It is worth noting that, as in most CRISPR associated RNases, nuclease activity at this site is metal-dependent, as target cleavage only occurs in the presence of a certain concentration of Mg2+ and is abolished after addition of EDTA.(5) This feature contrasts with most HEPN nucleases, where activity is metal-independent. A CRISPR associated RNase, Csx2 from Pyrococcus furious, contains a Zn2+ within its HEPN domain, suggesting a possible role of a divalent cation in further stabilizing the reaction intermediate (2). | + | HEPNs (Higher Eukaryotes and Prokaryotes Nucleotide-binding domain) are alpha helical domains, many of which are present in RNA maturation systems and related biological conflicts and most of which contain a catalytic Rx4-6H active site. Cas 13b <scene name='81/817991/Hepn1/1'>HEPN1</scene> domain is composed of 12 linearly connected alpha helices flexible loop between them, whereas <scene name='81/817991/Hepn2/1'>HEPN2</scene> contains nine alpha helices, several short beta strands and beta-hairpin with positively charged residues at the tip. In accordance with the conserved <scene name='81/817991/Hepn_siteb/1'>active site</scene> from HEPNs, a Rx4-6H site is present, where active residues locate within both HEPN1 (<scene name='81/817991/Hepn1_active_site/1'>R156, N157, H161</scene>) and HEPN2 (<scene name='81/817991/Hepn2_active_site/1'>R1068, N1069, H1073</scene>) domains. One of the two histidines is thought to act as a base, inducing the ribose 2’OH to attack the phosphodiester linkage. The conserved arginine could stabilize the negative oxygens of the transition state, as a similar residue (Lysine) has been shown to carry out such function in RNase A. Alternatively, it could interact with the RNA backbone. The other polar residues, located in between the catalytic histidine and the conserved arginine, are thought to further augment the active site. (2,4) It is worth noting that, as in most CRISPR associated RNases, nuclease activity at this site is metal-dependent, as target cleavage only occurs in the presence of a certain concentration of Mg2+ and is abolished after addition of EDTA.(5) This feature contrasts with most HEPN nucleases, where activity is metal-independent. A CRISPR associated RNase, Csx2 from Pyrococcus furious, contains a Zn2+ within its HEPN domain, suggesting a possible role of a divalent cation in further stabilizing the reaction intermediate (2). |
[[Image:Act1.png|550px|left|thumb| Fig.2 HEPN nuclease site]] | [[Image:Act1.png|550px|left|thumb| Fig.2 HEPN nuclease site]] | ||
Revision as of 12:12, 17 June 2019
==Cas13b==
| |||||||||||
References