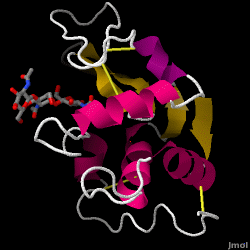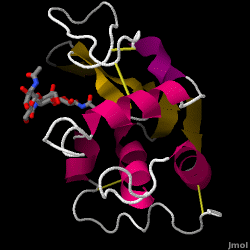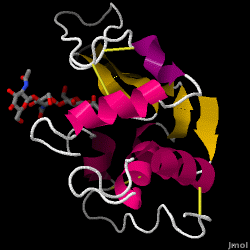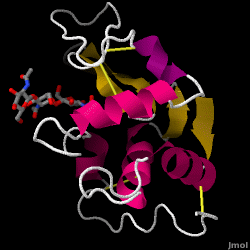
Hen Egg-White (HEW) Lysozyme
From Proteopedia
| Line 26: | Line 26: | ||
| - | <StructureSection load='' size='400' side='right' scene='37/376372/Overall/ | + | <StructureSection load='' size='400' side='right' scene='37/376372/Overall/3' caption='Hen egg white lysozyme (PDB code [[1hew]])'> |
== Structure of Lysozyme == | == Structure of Lysozyme == | ||
| - | <scene name='37/376372/Overall/ | + | <scene name='37/376372/Overall/3'>Lysozyme</scene> is a small single-subunit protein containing 129 amino acids. It folds into a compact structure with an active site cleft that binds to certain carbohydrates (<jmol><jmolLink><script> select ligand; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞⚟</text></jmolLink> </jmol>). Starting at the blue end, the N terminus(<jmol><jmolLink><script> select 1.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞⚟</text></jmolLink> </jmol>), one can work the entire way through to the C terminus (<jmol><jmolLink><script> select 129.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞⚟</text></jmolLink> </jmol>), showing the folding pattern of the protein. There are four disulfide bridges (<jmol><jmolLink><script> select BONDS ({0:3}); wireframe 0.6; delay 0.5; wireframe 0.1</script><text>⚞⚟</text></jmolLink> </jmol>) that crosslink the polypeptide, stabilizing the native conformation. Lysozyme's secondary structure comprises five alpha helices (<jmol><jmolLink><script> select helix and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞⚟</text></jmolLink> </jmol>)and five beta strands (<jmol><jmolLink><script> select sheet and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞⚟</text></jmolLink> </jmol>). Linking these, a number of beta turns and a large number of random coils make up the remainder of the polypeptide backbone. The secondary structure is easier to see in a <scene name='37/376372/Secondary/2'>ribbon view</scene>; however, this type of visualization (Richardson diagram) was not invented until about a quarter century after this first enzyme structure was solved, so the figures in the original report look more like the previous scene. |
=== Intermolecular Interactions === | === Intermolecular Interactions === | ||
'''Polarity''' | '''Polarity''' | ||
| - | The nature of the amino acid | + | The nature of the amino acid side chains in the lysozyme polypeptide sequence leads to regions of varying hydrophobicities and polarities of the enzyme structure. The presence of certain regions of hydrophilicity and hydrophobicity is a driving force in determining protein structure when folding. The varying polarities of the side chains influence the locations of residues in the enzyme structure. Nonpolar residues appear blue, and polar residues appear red in the following <scene name='Sandbox_38/Non_polar_blue/1'>polarity</scene> display of lysozyme. Nonpolar residues will display hydrophobic tendencies occurring mostly on the interior of the enzyme while polar residues will increase in abundance on the surface of the protein in order to increase contact with the aqueous solvent so as to satisfy their hydrophilic nature. By observing a space-filled structural depiction of <scene name='Sandbox_38/Non_polar_blu/1'>lysozyme polarity</scene> with polar molecules colored red and nonpolar molecules colored blue the influence of polarity on protein folding is evident, with the blue (nonpolar) regions inside the red (polar) regions. The presence of <scene name='Hen_Egg-White_(HEW)_Lysozyme/Wuter/1'>water</scene> interacting with the various hydrophilic residues is depicted to further display how polarity affects structure. Water is depicted as yellow, and the polar and nonpolar regions remain their respective color. |
| + | |||
| + | <jmol> | ||
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>hide (hidden and not sidechain) or (sidechain and not (met, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>hydrophobic side chains</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>hide (hidden and not sidechain) or (sidechain and (met, ile, leu, val, phe, tyr, trp))</script> | ||
| + | <text>hydrophilic side chains</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>display displayed or sidechain</script> | ||
| + | <text>all sidechains</text> | ||
| + | <checked>true</checked> | ||
| + | |||
| + | </item> | ||
| + | |||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>select sidechain; color gray; select sidechain and (met, ile, leu, val, phe, tyr, trp); color purple</script> | ||
| + | <text>color polar</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select sidechain; color gray; select sidechain and (asp, glu); color red; select sidechain and (lys, bluearg, his); color ; </script> | ||
| + | <text>color charge</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>display displayed or sidechain</script> | ||
| + | <text>all sidechains</text> | ||
| + | <checked>true</checked> | ||
| + | |||
| + | </item> | ||
| + | |||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | |||
| + | |||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>display displayed or water</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>hide water or hidden</scriptWhenUnchecked> | ||
| + | <checked>true</checked> | ||
| + | <text>water</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol> | ||
'''Hydrogen Bonding''' | '''Hydrogen Bonding''' | ||
Revision as of 19:33, 20 June 2019
Lysozyme - also known as muramidase, or glycoside hydrolase - is a powerful enzyme of biological significance found in abundance in tears, saliva, and human milk. In humans, it is encoded in the LYZ gene. Although it is responsible for the initial digestion of starches in the mouth, it is most widely identified as a non-specific defense in gram positive bacteria and in many species of fungi. Due to its antibacterial effects, it is a strong component of the innate immune system, and is an important part of an infant's diet to ward off diarrheal diseases. Since it is a small, easily available, and highly stable protein containing only 129 amino acid residues, it has been subject to extensive research regarding its function and structure. Hen Egg White (HEW) Lysozyme is shown below.
Introduction
Lysozyme is an enzyme known for its unique ability to degrade the polysaccharide architecture of many kinds of cell walls, normally for the purpose of protection against bacterial infection[1]. Its effects were first noticed by Laschtschenko in 1909. It was officially characterized and termed “lysozyme” by Alexander Fleming, the same person credited for the accidental discovery of penicillin.
The characterization of lysozyme in 1922 by Alexander Fleming was providential in that the undertaken experiment related to the discovery of lysozyme was not geared toward any knowledge of such a protein as lysozyme [2]. During the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." Fleming's discovery was complemented by David C. Phillips' 1965 description of the three-dimensional structure of lysozyme via a 200 pm resolution model obtained from X-ray crystallography [3]. Phillips' work was especially groundbreaking since Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished[4]. Phillips' research also led to the first sufficiently described enzymatic mechanism of catalytic action [5]. Thus, Phillips' elucidation of the function of lysozyme led Phillips to reach a more general conclusion on the diversity of enzymatic chemical action in relation to enzymatic structure. Clearly, the findings of Phillips as well as the more general historical development of the understanding of the structure and function of lysozyme have been paramount to the more general realm of enzyme chemistry.
See also
Function
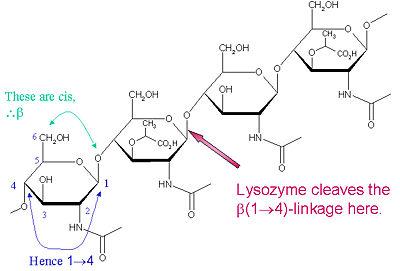
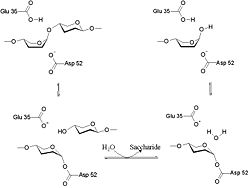
Lysozyme is known for damaging bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid (NAM) and N-acetyl-D-glucosamine (NAG) residues in peptidoglycan, and between N-acetyl-D-glucosamine residues in chitodextrins. In this way, lysozyme is efficient in lysing the cell walls of both bacteria and fungi. The location of cleavage for lysozyme on this architectural theme is the β(1-4) glycosidic linkage connecting the C1 carbon of NAM to the C4 carbon of NAG.
The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the cleaving target glycosidic bond, NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cleavage preference.
| |||||||||||
See Also
References
- ↑ Lysozyme. 2010. Citizendium.org. http://en.citizendium.org/wiki/Lysozyme
- ↑ Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford
- ↑ 1967. Proc R Soc Lond B Bio 167 (1009): 389–401.
- ↑ Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm
- ↑ http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm
- ↑ http://www.worthington-biochem.com/ly/default.html
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Johnson LN, Phillips DC. Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution. Nature. 1965 May 22;206(986):761-3. PMID:5840126
- ↑ Phillips, D. C. The hen egg white lysozyme molecule. Proc. Natl Acad. Sci. USA 57, 483-495 (1967)
- ↑ coordinates of the model kindly provided by Louise Johnson
- ↑ Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001 Aug 23;412(6849):835-8. PMID:11518970 doi:10.1038/35090602
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Anne Goodling, Alexander Berchansky, Eric Martz, Joel L. Sussman, Karl Oberholser, Jaime Prilusky, John Ripollone, Daniel Kreider, Judy Voet, John S. de Banzie