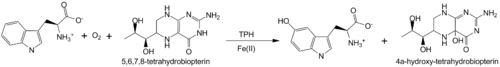We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hydroxylase
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
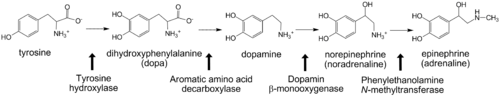
Aromatic amino acid hydroxylases consists of phenylalanine hydroxylase, tyrosine hydroxylase and tryptophan hydroxylase. These enzymes all contain iron and use BH<sub>4</sub> as a co-substrate in the hydroxylation of their respective aromatic amino acids. Additionally all mammalian AAAH form homotetramers and each monomer consists of three domains. These domains are the N-terminal regulatory domain (100-150 residues), the catalytic domain (approximately 315 residues) and the C-terminal tetramerization domain (approximately 30-40 residues)<ref>PMID: 10800597</ref>. | Aromatic amino acid hydroxylases consists of phenylalanine hydroxylase, tyrosine hydroxylase and tryptophan hydroxylase. These enzymes all contain iron and use BH<sub>4</sub> as a co-substrate in the hydroxylation of their respective aromatic amino acids. Additionally all mammalian AAAH form homotetramers and each monomer consists of three domains. These domains are the N-terminal regulatory domain (100-150 residues), the catalytic domain (approximately 315 residues) and the C-terminal tetramerization domain (approximately 30-40 residues)<ref>PMID: 10800597</ref>. | ||
| - | '''Phenylalanine hydroxylase''' (PAH) is found in the liver where it catalyses the hydroxylation of phenylalanine to tyrosine. This is the first step in the oxidative degradation of phenylalanine<ref>PMID: 13428782</ref>. Mutations in PAH leading to a decrease in enzyme activity result in the disease phenylketonuria, where phenylalanine is converted to phenylpyruvate. Phenylpyruvate is toxic and leads to mental retardation. PAH is also found in some bacteria, but only PAH from the bacteria ''Chromobacterium violaceum''<ref>PMID: 9748224</ref> and ''Colwellia psychrerythraea''<ref>PMID: 17537732</ref> have been characterized. The bacterial PAHs are monomeric and do not contain a regulatory domain. All the | + | '''Phenylalanine hydroxylase''' (PAH) is found in the liver where it catalyses the hydroxylation of phenylalanine to tyrosine. This is the first step in the oxidative degradation of phenylalanine<ref>PMID: 13428782</ref>. Mutations in PAH leading to a decrease in enzyme activity result in the disease phenylketonuria, where phenylalanine is converted to phenylpyruvate. Phenylpyruvate is toxic and leads to mental retardation. PAH is also found in some bacteria, but only PAH from the bacteria ''Chromobacterium violaceum''<ref>PMID: 9748224</ref> and ''Colwellia psychrerythraea''<ref>PMID: 17537732</ref> have been characterized. The bacterial PAHs are monomeric and do not contain a regulatory domain. All the aromatic amino acid hydroxylases are believed to have evolved from common ancestor containing only the catalytic domain<ref>PMID: 3475690</ref>. |
<br/> | <br/> | ||
See also [[Protein Stability and in Vivo Concentration of Missense Mutations in Phenylalanine Hydroxylase]]. | See also [[Protein Stability and in Vivo Concentration of Missense Mutations in Phenylalanine Hydroxylase]]. | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
References
- ↑ Fitzpatrick PF. The aromatic amino acid hydroxylases. Adv Enzymol Relat Areas Mol Biol. 2000;74:235-94. PMID:10800597
- ↑ KAUFMAN S. The enzymatic conversion of phenylalanine to tyrosine. J Biol Chem. 1957 May;226(1):511-24. PMID:13428782
- ↑ Chen D, Frey PA. Phenylalanine hydroxylase from Chromobacterium violaceum. Uncoupled oxidation of tetrahydropterin and the role of iron in hyroxylation. J Biol Chem. 1998 Oct 2;273(40):25594-601. PMID:9748224
- ↑ Leiros HK, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J Biol Chem. 2007 Jul 27;282(30):21973-86. Epub 2007 May 30. PMID:17537732 doi:10.1074/jbc.M610174200
- ↑ Grenett HE, Ledley FD, Reed LL, Woo SL. Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5530-4. PMID:3475690
- ↑ NAGATSU T, LEVITT M, UDENFRIEND S. TYROSINE HYDROXYLASE. THE INITIAL STEP IN NOREPINEPHRINE BIOSYNTHESIS. J Biol Chem. 1964 Sep;239:2910-7. PMID:14216443
- ↑ 7.0 7.1 LEVINE RJ, LOVENBERG W, SJOERDSMA A. HYDROXYLATION OF TRYPTOPHAN AND PHENYLALANINE IN NEOPLASTIC MAST CELLS OF THE MOUSE. Biochem Pharmacol. 1964 Sep;13:1283-90. PMID:14221726
- ↑ Grahame-Smith DG. Tryptophan hydroxylation in brain. Biochem Biophys Res Commun. 1964 Aug 11;16(6):586-92. PMID:5297063
- ↑ 9.0 9.1 Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochem Pharmacol. 2003 Nov 1;66(9):1673-80. PMID:14563478
- ↑ Grenett HE, Ledley FD, Reed LL, Woo SL. Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5530-4. PMID:3475690
- ↑ Craig SP, Boularand S, Darmon MC, Mallet J, Craig IW. Localization of human tryptophan hydroxylase (TPH) to chromosome 11p15.3----p14 by in situ hybridization. Cytogenet Cell Genet. 1991;56(3-4):157-9. PMID:2055111
- ↑ Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003 Jan 3;299(5603):76. PMID:12511643 doi:http://dx.doi.org/10.1126/science.1078197
- ↑ 13.0 13.1 Patel PD, Pontrello C, Burke S. Robust and tissue-specific expression of TPH2 versus TPH1 in rat raphe and pineal gland. Biol Psychiatry. 2004 Feb 15;55(4):428-33. PMID:14960297 doi:http://dx.doi.org/10.1016/j.biopsych.2003.09.002
- ↑ Slominski A, Pisarchik A, Johansson O, Jing C, Semak I, Slugocki G, Wortsman J. Tryptophan hydroxylase expression in human skin cells. Biochim Biophys Acta. 2003 Oct 15;1639(2):80-6. PMID:14559114
- ↑ Hasegawa H, Yanagisawa M, Inoue F, Yanaihara N, Ichiyama A. Demonstration of non-neural tryptophan 5-mono-oxygenase in mouse intestinal mucosa. Biochem J. 1987 Dec 1;248(2):501-9. PMID:3435461
- ↑ Hosoda S, Nakamura W, Takatsuki K. Properties of tryptophan hydroxylase from human carcinoid tumor. Biochim Biophys Acta. 1977 May 12;482(1):27-34. PMID:16654
- ↑ Manuck SB, Flory JD, Ferrell RE, Dent KM, Mann JJ, Muldoon MF. Aggression and anger-related traits associated with a polymorphism of the tryptophan hydroxylase gene. Biol Psychiatry. 1999 Mar 1;45(5):603-14. PMID:10088047
- ↑ Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med. 2011 Aug;13(8):697-707. doi: 10.1097/GIM.0b013e3182141b48. PMID:21555948 doi:http://dx.doi.org/10.1097/GIM.0b013e3182141b48
- ↑ Flatmark T, Stevens RC. Structural Insight into the Aromatic Amino Acid Hydroxylases and Their Disease-Related Mutant Forms. Chem Rev. 1999 Aug 11;99(8):2137-2160. PMID:11849022
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Michael Skovbo Windahl, David Canner, Joel L. Sussman, Jaime Prilusky