RNA
From Proteopedia
| Line 12: | Line 12: | ||
===Genetic information=== | ===Genetic information=== | ||
RNA plays key roles in gene expression, in the form of mRNA (messenger RNA), tRNA (transfer RNA) and rRNA (ribosomal RNA), the classic three types of RNA. Beyond these roles, RNA also serves to store genetic information and to regulate and modulate it. | RNA plays key roles in gene expression, in the form of mRNA (messenger RNA), tRNA (transfer RNA) and rRNA (ribosomal RNA), the classic three types of RNA. Beyond these roles, RNA also serves to store genetic information and to regulate and modulate it. | ||
| - | |||
| - | ====Storage==== | ||
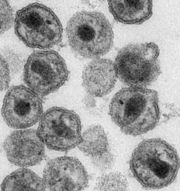
| - | [[Image:HIV_particles.JPG|thumb]]All known organisms use DNA as the storage molecule for genetic information. Some viruses, however use RNA instead. They are classified into double-stranded RNA viruses and plus or minus single-stranded RNA viruses depending on how they use RNA to store genetic information. For example, human immunodeficiency virus carries genetic information in the form of single stranded RNA. In the figure, the RNA is located in the dark rectangular blob inside the circular virus particles. | ||
====Transcription==== | ====Transcription==== | ||
| Line 24: | Line 21: | ||
====Translation==== | ====Translation==== | ||
Ribosomes synthesize proteins with sequences determined by mRNA. The ribosome is a complex of rRNA and protein. Shown here is a small part of the ribosome illustrating protein-RNA interactions: <scene name='49/491851/Ribosomal/1'>ribosomal RNA</scene> (<jmol><jmolLink><script> select rna; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞RNA⚟</text></jmolLink> </jmol>). When synthesizing proteins, ribosomes follow the genetic code to translate an RNA sequence (grouped into codons of 3 nucleotides) into a protein sequence.<scene name='43/433638/Wireframe/5'>tRNAs</scene> are the adapter molecules that embody the genetic code. Each tRNA is covalently bound to a specific amino acid at one end and display an anti-codon on the other end (for more details, see [[transfer RNA]]) | Ribosomes synthesize proteins with sequences determined by mRNA. The ribosome is a complex of rRNA and protein. Shown here is a small part of the ribosome illustrating protein-RNA interactions: <scene name='49/491851/Ribosomal/1'>ribosomal RNA</scene> (<jmol><jmolLink><script> select rna; selectionHalos ON; delay 0.5;selectionHalos OFF;</script><text>⚞RNA⚟</text></jmolLink> </jmol>). When synthesizing proteins, ribosomes follow the genetic code to translate an RNA sequence (grouped into codons of 3 nucleotides) into a protein sequence.<scene name='43/433638/Wireframe/5'>tRNAs</scene> are the adapter molecules that embody the genetic code. Each tRNA is covalently bound to a specific amino acid at one end and display an anti-codon on the other end (for more details, see [[transfer RNA]]) | ||
| + | |||
| + | ====Storage of genetic information==== | ||
| + | [[Image:HIV_particles.JPG|thumb]]All known organisms store genetic information on DNA. Some viruses, however, use RNA instead. Depending on how they use RNA to store genetic information, they are classified as double-stranded RNA viruses and plus or minus single-stranded RNA viruses . For example, human immunodeficiency virus carries genetic information in the form of single stranded RNA. In the figure, the RNA is located in the dark rectangular blob inside the circular virus particles. The single-stranded RNA folds onto itself, as illustrated for the part of the RNA called <scene name='49/491851/Hiv_rna/1'>packaging signal</scene>. | ||
| + | |||
===Catalysis=== | ===Catalysis=== | ||
Revision as of 22:24, 1 January 2022
RNA (ribonucleic acid) is a biological macromolecule that stores and processes genetic information, catalyzes chemical reactions and regulates biological processes. Just like the other nucleic acid (DNA), it is a linear polymer (a "strand") of nucleotide building blocks. These building blocks themselves are made up of a sugar linked to a (nitrogenous) base and a phosphate. RNA utilizes a different set of bases than DNA, and it sugar (ribose) contains one additional hydroxyl group compared to that of DNA (deoxyribose). Different than DNA, which mostly occurs in pairs of complementary strands in its biological context, RNA mostly lacks a complementary strand, leading to a larger variety of 3D structures and biological functions.
RNA structure and function
| |||||||||||
Chemistry
- Base: flat, aromatic, hydrogen-bonding capability, base modifications, base pairing, base stacking
- Sugar phosphate backbone: torsion angles, sugar pucker, phosphate-phosphate distance
Transcription and Translation
The expression of genes into proteins and is a process involving two stages called transcription and translation. In the transcription stage a strand of DNA molecule serves as a template for the synthesis of an RNA molecule called messenger RNA. In the case of RNAs that code for polypetides, this messenger RNA is then translated into proteins on ribosomes.
Post-transcriptionally, specific nucleotides in RNA are often further modified. This is most frequent in [tRNA|transfer ribonucleic acid (tRNAs)]], the adapter molecules of Translation, and ribosomal ribonucleic acids (rRNAs) of the ribosome.
See Also
- Base stacking
- Ribosome
- Translation
- DNA Replication, Transcription and Translation
- Transfer ribonucleic acid (tRNA)
- Ribozyme
- Kink-turn motif
- Pseudouridine
- Non-Standard Residues
- DNA
- For additional information, see: Nucleic Acids
External Resources
- HD-RNAS: Hierarchical Database of RNA Structures is a systematic hierarchical organization classifying all RNAs available in the Protein Data Bank. This helps make it easier to navigate the availble structural data given the large numbers of redundant files and ambiguous synthetic sequences.
- Liley Database
References
- ↑ Steitz TA. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr Opin Struct Biol. 2009 Dec;19(6):683-90. doi: 10.1016/j.sbi.2009.09.001., Epub 2009 Oct 5. PMID:19811903 doi:http://dx.doi.org/10.1016/j.sbi.2009.09.001
- ↑ Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. 2008 Nov;15(11):1221-2. Epub 2008 Oct 26. PMID:18953333 doi:10.1038/nsmb.1509