We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1hzy
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
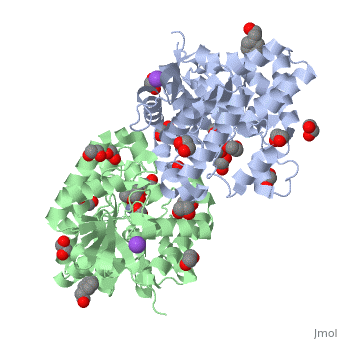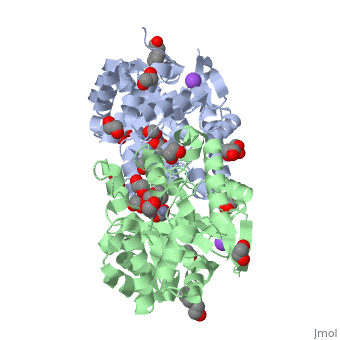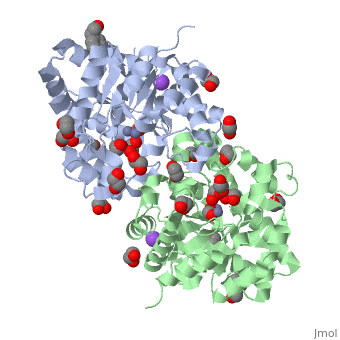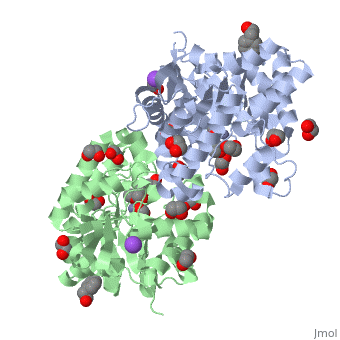
<StructureSection load='1hzy' size='340' side='right'caption='[[1hzy]], [[Resolution|resolution]] 1.30Å' scene=''> | <StructureSection load='1hzy' size='340' side='right'caption='[[1hzy]], [[Resolution|resolution]] 1.30Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1hzy]] is a 2 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1hzy]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Aj_2067 Aj 2067]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1HZY OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1HZY FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=FMT:FORMIC+ACID'>FMT</scene>, <scene name='pdbligand=NA:SODIUM+ION'>NA</scene>, <scene name='pdbligand=PEL:2-PHENYL-ETHANOL'>PEL</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=FMT:FORMIC+ACID'>FMT</scene>, <scene name='pdbligand=NA:SODIUM+ION'>NA</scene>, <scene name='pdbligand=PEL:2-PHENYL-ETHANOL'>PEL</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1i03|1i03]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[1i03|1i03]]</div></td></tr> |
| - | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[https://en.wikipedia.org/wiki/Aryldialkylphosphatase Aryldialkylphosphatase], with EC number [https://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.1.8.1 3.1.8.1] </span></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1hzy FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1hzy OCA], [https://pdbe.org/1hzy PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1hzy RCSB], [https://www.ebi.ac.uk/pdbsum/1hzy PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1hzy ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/OPD_BREDI OPD_BREDI]] Has an unusual substrate specificity for synthetic organophosphate triesters and phosphorofluoridates. All of the phosphate triesters found to be substrates are synthetic compounds. The identity of any naturally occurring substrate for the enzyme is unknown. Has no detectable activity with phosphate monoesters or diesters and no activity as an esterase or protease. It catalyzes the hydrolysis of the insecticide paraoxon at a rate approaching the diffusion limit and thus appears to be optimally evolved for utilizing this synthetic substrate. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 32: | Line 32: | ||
==See Also== | ==See Also== | ||
| - | + | *[[Phosphotriesterase 3D structures|Phosphotriesterase 3D structures]] | |
| - | *[[Phosphotriesterase|Phosphotriesterase]] | + | |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 10:55, 4 August 2021
HIGH RESOLUTION STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA
| |||||||||||
Categories: Aj 2067 | Aryldialkylphosphatase | Large Structures | Benning, M M | Holden, H M | Raushel, F M | Shim, H | Hydrolase | Pte | Zinc