We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hemeproteins
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
=== ''Rhodothermus marinus'' cytochrome ''c'' === | === ''Rhodothermus marinus'' cytochrome ''c'' === | ||
| - | + | '''Structure''' | |
<scene name='Sandbox_Reserved_335/Heme/1'>'Figure 1. The heme group of monoheme cytochrome ''c'' purified from ''Rhodothermus marinus''</scene> | <scene name='Sandbox_Reserved_335/Heme/1'>'Figure 1. The heme group of monoheme cytochrome ''c'' purified from ''Rhodothermus marinus''</scene> | ||
| Line 54: | Line 54: | ||
The observation of these structural motifs in other C-type cytochromes can support the divergent evolution of cytochromes ''c''.<ref name=main /> These motifs are present in a number of different bacteria and are seen in similar regions of the secondary structure; however, they exist in the primary sequence in places distinct to the phylum. For example, monoheme cytochromes ''c'' in the rest of the Bacteroidetes phylum have an N-terminus extension that is highly conserved to that of ''Rm''cyt''c'', and the regions in the primary structure that correspond to these secondary motifs are not observed in other bacterial phyla.<ref name=main /> Also, due to these motifs being absent from other phyla, the Bacteroidetes monoheme cyt ''c'' has been said to form a new subfamily of cyt ''c''. | The observation of these structural motifs in other C-type cytochromes can support the divergent evolution of cytochromes ''c''.<ref name=main /> These motifs are present in a number of different bacteria and are seen in similar regions of the secondary structure; however, they exist in the primary sequence in places distinct to the phylum. For example, monoheme cytochromes ''c'' in the rest of the Bacteroidetes phylum have an N-terminus extension that is highly conserved to that of ''Rm''cyt''c'', and the regions in the primary structure that correspond to these secondary motifs are not observed in other bacterial phyla.<ref name=main /> Also, due to these motifs being absent from other phyla, the Bacteroidetes monoheme cyt ''c'' has been said to form a new subfamily of cyt ''c''. | ||
| - | + | '''Function''' | |
Monoheme cytochromes ''c'' are involved in electron transport chains in both prokaryotes and eukaryotic mitochondria.<ref name=main /> They mediate the transfer of electrons mainly from the ''bc''<sub>1</sub> complexes or their analogs to heme-copper oxygen reductases (HCOs) in the [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain] of [http://en.wikipedia.org/wiki/Oxidative_phosphorylation oxidative phosphorylation]. Heme ''c'' containing domains are often found fused to other protein domains such as these HCOs, including the ''caa''<sub>3</sub> oxygen reductases<ref name=main /><ref>PMID:14691678</ref>; these enzymes are membrane-bound and catalyze the reduction of O<sub>2</sub> to water.<ref>PMID:11334784</ref> In addition to being involved in oxidative phosphorylation, monoheme cyt ''c'' has also been seen to participate in the electron transport chain of [http://en.wikipedia.org/wiki/Photosynthesis photosynthesis].<ref name=main /> Cytochrome ''c'' has also been determined to be a major signalling molecule in the apoptotic pathways. | Monoheme cytochromes ''c'' are involved in electron transport chains in both prokaryotes and eukaryotic mitochondria.<ref name=main /> They mediate the transfer of electrons mainly from the ''bc''<sub>1</sub> complexes or their analogs to heme-copper oxygen reductases (HCOs) in the [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain] of [http://en.wikipedia.org/wiki/Oxidative_phosphorylation oxidative phosphorylation]. Heme ''c'' containing domains are often found fused to other protein domains such as these HCOs, including the ''caa''<sub>3</sub> oxygen reductases<ref name=main /><ref>PMID:14691678</ref>; these enzymes are membrane-bound and catalyze the reduction of O<sub>2</sub> to water.<ref>PMID:11334784</ref> In addition to being involved in oxidative phosphorylation, monoheme cyt ''c'' has also been seen to participate in the electron transport chain of [http://en.wikipedia.org/wiki/Photosynthesis photosynthesis].<ref name=main /> Cytochrome ''c'' has also been determined to be a major signalling molecule in the apoptotic pathways. | ||
| - | + | '''Electron transport chain''' | |
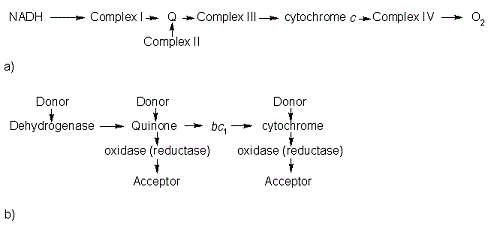
In the electron transport chain (ETC), cyt ''c'' shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome ''bc''<sub>1</sub> complex and IV is cyt ''c'' oxidase. Initially, the heme iron in cyt ''c'' is in the reduced, Fe<sup>3+</sup> state; this allows for the uptake of one electron, oxidizing the iron to the Fe<sup>2+</sup> state.<ref name='etc'>Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.</ref> The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3). | In the electron transport chain (ETC), cyt ''c'' shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome ''bc''<sub>1</sub> complex and IV is cyt ''c'' oxidase. Initially, the heme iron in cyt ''c'' is in the reduced, Fe<sup>3+</sup> state; this allows for the uptake of one electron, oxidizing the iron to the Fe<sup>2+</sup> state.<ref name='etc'>Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.</ref> The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3). | ||
| Line 67: | Line 67: | ||
The cytochrome oxidase reaction accounts for nearly 90% of all oxygen uptake in most cells.<ref name=etc /> Due to the large role of cytochromes within the ETC, it would be highly detrimental to the cell if any inhibitors were to be present in the organism. Cyanide and azide bind tightly to the cytochrome oxidase complex, halting electron transport and reducing the overall ATP production.<ref name=etc /> | The cytochrome oxidase reaction accounts for nearly 90% of all oxygen uptake in most cells.<ref name=etc /> Due to the large role of cytochromes within the ETC, it would be highly detrimental to the cell if any inhibitors were to be present in the organism. Cyanide and azide bind tightly to the cytochrome oxidase complex, halting electron transport and reducing the overall ATP production.<ref name=etc /> | ||
| - | + | '''Apoptosis''' | |
In all organisms, cells undergo [http://en.wikipedia.org/wiki/Apoptosis apoptosis], or programmed cell death, by which there is an extrinsic and an intrinsic pathway. The extrinsic pathway involves an immune response by killer lymphocytes, and once the lymphocyte has been bound to the target cell, an apoptotic cascade occurs.<ref name=etc /> The intrinsic pathway includes cyt ''c'', present in the intermembrane space of mitochondria. In this pathway, the presence of an apoptotic stimulus causes cyt ''c'' to be released into the cytosol. Cytochrome ''c'' in the cytosol now can be recognized and bound to various apoptotic factors, activating them and forming the [http://en.wikipedia.org/wiki/Apoptosome apoptosome]. The apoptosome recruits [http://en.wikipedia.org/wiki/Caspase caspases], which are activated and result in a caspase cascade to proceed with apoptosis.<ref name=etc /> | In all organisms, cells undergo [http://en.wikipedia.org/wiki/Apoptosis apoptosis], or programmed cell death, by which there is an extrinsic and an intrinsic pathway. The extrinsic pathway involves an immune response by killer lymphocytes, and once the lymphocyte has been bound to the target cell, an apoptotic cascade occurs.<ref name=etc /> The intrinsic pathway includes cyt ''c'', present in the intermembrane space of mitochondria. In this pathway, the presence of an apoptotic stimulus causes cyt ''c'' to be released into the cytosol. Cytochrome ''c'' in the cytosol now can be recognized and bound to various apoptotic factors, activating them and forming the [http://en.wikipedia.org/wiki/Apoptosome apoptosome]. The apoptosome recruits [http://en.wikipedia.org/wiki/Caspase caspases], which are activated and result in a caspase cascade to proceed with apoptosis.<ref name=etc /> | ||
Revision as of 13:34, 3 November 2019
| |||||||||||
References
- ↑ Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003 Feb;97(2):139-52. PMID:12559387
- ↑ Rodriguez-Maranon MJ, Qiu F, Stark RE, White SP, Zhang X, Foundling SI, Rodriguez V, Schilling CL 3rd, Bunce RA, Rivera M. 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c. Biochemistry. 1996 Dec 17;35(50):16378-90. PMID:8973214 doi:10.1021/bi961895o
- ↑ Crofts AR. The cytochrome bc1 complex: function in the context of structure. Annu Rev Physiol. 2004;66:689-733. PMID:14977419 doi:http://dx.doi.org/10.1146/annurev.physiol.66.032102.150251
- ↑ Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-Ray Structure of Rhodobacter Capsulatus Cytochrome bc (1): Comparison with its Mitochondrial and Chloroplast Counterparts. Photosynth Res. 2004;81(3):251-75. PMID:16034531 doi:http://dx.doi.org/10.1023/B:PRES.0000036888.18223.0e
- ↑ Rajagopal BS, Wilson MT, Bendall DS, Howe CJ, Worrall JA. Structural and kinetic studies of imidazole binding to two members of the cytochrome c (6) family reveal an important role for a conserved heme pocket residue. J Biol Inorg Chem. 2011 Jan 26. PMID:21267610 doi:10.1007/s00775-011-0758-y
- ↑ Morelli X, Czjzek M, Hatchikian CE, Bornet O, Fontecilla-Camps JC, Palma NP, Moura JJ, Guerlesquin F. Structural model of the Fe-hydrogenase/cytochrome c553 complex combining transverse relaxation-optimized spectroscopy experiments and soft docking calculations. J Biol Chem. 2000 Jul 28;275(30):23204-10. PMID:10748163 doi:10.1074/jbc.M909835199
- ↑ Manole A, Kekilli D, Svistunenko DA, Wilson MT, Dobbin PS, Hough MA. Conformational control of the binding of diatomic gases to cytochrome c'. J Biol Inorg Chem. 2015 Mar 20. PMID:25792378 doi:http://dx.doi.org/10.1007/s00775-015-1253-7
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 Stelter M, Melo AM, Pereira MM, Gomes CM, Hreggvidsson GO, Hjorleifsdottir S, Saraiva LM, Teixeira M, Archer M. A Novel Type of Monoheme Cytochrome c: Biochemical and Structural Characterization at 1.23 A Resolution of Rhodothermus marinus Cytochrome c. Biochemistry. 2008 Oct 15. PMID:18855424 doi:10.1021/bi800999g
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedheme - ↑ Than ME, Hof P, Huber R, Bourenkov GP, Bartunik HD, Buse G, Soulimane T. Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing. J Mol Biol. 1997 Aug 29;271(4):629-44. PMID:9281430 doi:10.1006/jmbi.1997.1181
- ↑ Soares CM, Baptista AM, Pereira MM, Teixeira M. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J Biol Inorg Chem. 2004 Mar;9(2):124-34. Epub 2003 Dec 23. PMID:14691678 doi:10.1007/s00775-003-0509-9
- ↑ Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001 Jun 1;1505(2-3):185-208. PMID:11334784
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.