We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Succinate Dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
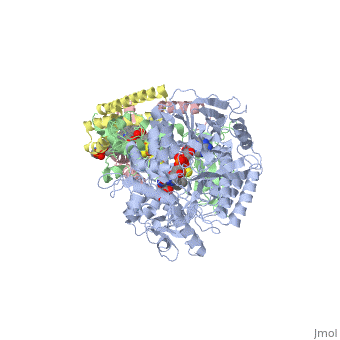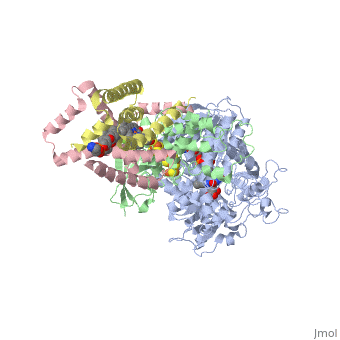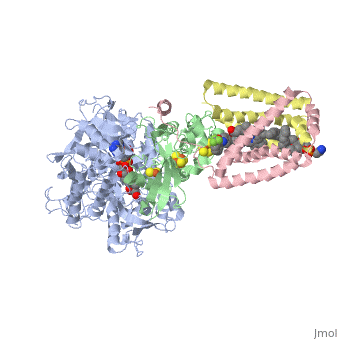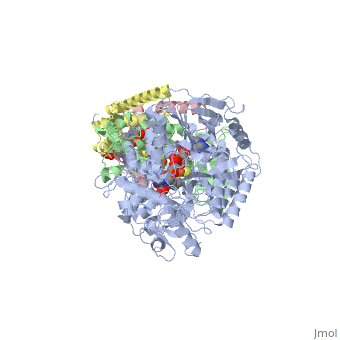
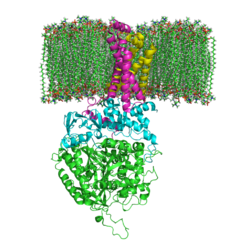
<StructureSection load='3ae1' size='350' side='right' caption='Succinate dehydrogenase containing flavoprotein subunit (Sdha) (grey), iron-sulfur subunit (Sdhb) (green), cytochrome B560 subunit (Sdhc) (pink) and cytochrome B small subunit (Sdhd) (yellow), FAD, Fe2S2, Fe4S4 and Fe3S4 complex with protoporphyrin, malonate, benzamide derivative and α-phosphatidyl-β-oleoyl-γ-palmitoyl-phosphatidylethanolamine, [[3ae1]] ' scene=''> | <StructureSection load='3ae1' size='350' side='right' caption='Succinate dehydrogenase containing flavoprotein subunit (Sdha) (grey), iron-sulfur subunit (Sdhb) (green), cytochrome B560 subunit (Sdhc) (pink) and cytochrome B small subunit (Sdhd) (yellow), FAD, Fe2S2, Fe4S4 and Fe3S4 complex with protoporphyrin, malonate, benzamide derivative and α-phosphatidyl-β-oleoyl-γ-palmitoyl-phosphatidylethanolamine, [[3ae1]] ' scene=''> | ||
[[Image:Succinate Dehydrogenase.png|left|250px]] | [[Image:Succinate Dehydrogenase.png|left|250px]] | ||
| - | + | {{Clear}} | |
== Function == | == Function == | ||
[[Succinate Dehydrogenase]] (PDB = [[2wdv]] with empty ubiquinone binding site; PDB = [[1nek]] with ubiquinone bound), also called '''succinate-coenzyme Q reductase''' (SQR) or '''Complex II''', is a tetrameric enzyme found in the cell membrane of http://proteopedia.org/wiki/index.php?title=Succinate_Dehydrogenase&action=editsome bacteria and the inner mitochondrial membrane of mammalian cells. It is classified as an α+β protein, as it contains <scene name='Michael_Vick_Sandbox_2/2wdv_sec_structure/1'>segregated regions</scene> of α helices and antiparallel β sheets. It is involved in two aspects of digestion; it catalyzes the oxidation of succinate to fumarate in the [[The_Citric_Acid_Cycle|citric acid cycle]] by simultaneously reducing ubiquinone to ubiquinol in the electron transport chain <ref>PMID:14672929</ref>. See also:<br /> | [[Succinate Dehydrogenase]] (PDB = [[2wdv]] with empty ubiquinone binding site; PDB = [[1nek]] with ubiquinone bound), also called '''succinate-coenzyme Q reductase''' (SQR) or '''Complex II''', is a tetrameric enzyme found in the cell membrane of http://proteopedia.org/wiki/index.php?title=Succinate_Dehydrogenase&action=editsome bacteria and the inner mitochondrial membrane of mammalian cells. It is classified as an α+β protein, as it contains <scene name='Michael_Vick_Sandbox_2/2wdv_sec_structure/1'>segregated regions</scene> of α helices and antiparallel β sheets. It is involved in two aspects of digestion; it catalyzes the oxidation of succinate to fumarate in the [[The_Citric_Acid_Cycle|citric acid cycle]] by simultaneously reducing ubiquinone to ubiquinol in the electron transport chain <ref>PMID:14672929</ref>. See also:<br /> | ||
Revision as of 13:45, 7 November 2019
| |||||||||||
3D structures of succinate dehydrogenase
Updated on 07-November-2019
References
- ↑ Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004 Mar 5;279(10):9424-31. Epub 2003 Dec 12. PMID:14672929 doi:10.1074/jbc.M311876200
- ↑ Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K. Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase). J Biochem. 2003 Aug;134(2):191-5. PMID:12966066
- ↑ 3.0 3.1 Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003 Jan 31;299(5607):700-4. PMID:12560550 doi:10.1126/science.1079605
- ↑ 4.0 4.1 Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006 Mar 17;281(11):7309-16. Epub 2005 Dec 27. PMID:16407191 doi:http://dx.doi.org/10.1074/jbc.M508173200
- ↑ Kenney WC. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975 Apr 25;250(8):3089-94. PMID:235539
- ↑ Voet, Donald, Charlotte W. Pratt, and Judith G. Voet. Fundamentals of Biochemistry: Life at the Molecular Level. 2nd Ed. Hoboken, NJ: Wiley, 2008.
- ↑ Vinogradov AD, Kotlyar AB, Burov VI, Belikova YO. Regulation of succinate dehydrogenase and tautomerization of oxaloacetate. Adv Enzyme Regul. 1989;28:271-80. PMID:2624174
- ↑ Boyd AW, Lackmann M. Signals from Eph and ephrin proteins: a developmental tool kit. Sci STKE. 2001 Dec 11;2001(112):re20. PMID:11741094 doi:10.1126/stke.2001.112.re20
- ↑ 9.0 9.1 9.2 Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200
- ↑ Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, Van Remmen H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem J. 2008 Jan 15;409(2):491-9. PMID:17916065 doi:10.1042/BJ20071162
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Michael Vick, David Canner, Alexander Berchansky