We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1559
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
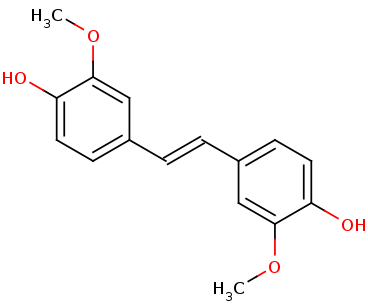
The flat surface of the B-sheet is pushing the amino acids up, making it possible for the <scene name='82/823083/6ojttriad/1'>catalytic triad</scene> Phe59, Tyr101, and Lys134 to create interactions with the <scene name='82/823083/Nsl_ligand/1'>ligand</scene>. | The flat surface of the B-sheet is pushing the amino acids up, making it possible for the <scene name='82/823083/6ojttriad/1'>catalytic triad</scene> Phe59, Tyr101, and Lys134 to create interactions with the <scene name='82/823083/Nsl_ligand/1'>ligand</scene>. | ||
| - | + | ||
The tertiary structure creates a<scene name='82/823083/Aminobindingpocket/1'> binding pocket of amino acids</scene> that are important to the active site. His282 provides pi-stacking, Phe305 provides Hydrophobic contacts, and Tyr101 provides Hydrogen bonding. The tertiary structure also allows the NSL ligand to interact using its 4-hydroxy with the catalytic triad. LsdA can only cleave 4-hydroxystilbenes. The photo below shows the NSL ligand interacting in the binding pocket. | The tertiary structure creates a<scene name='82/823083/Aminobindingpocket/1'> binding pocket of amino acids</scene> that are important to the active site. His282 provides pi-stacking, Phe305 provides Hydrophobic contacts, and Tyr101 provides Hydrogen bonding. The tertiary structure also allows the NSL ligand to interact using its 4-hydroxy with the catalytic triad. LsdA can only cleave 4-hydroxystilbenes. The photo below shows the NSL ligand interacting in the binding pocket. | ||
| - | + | ||
The tertiary structure also creates a metal binding site of Histidines to keep the iron molecule in place. There have been two mechanisms proposed for Lsd's. In one mechanism, the hydroxystillbenoid is activated via the enzyme-catalyzed deprotonation of the 4-hydroxy group, which then allows electron delocalization toward an Fe3+. In the other mechanism, π electron density from the double bond is redistributed to the iron-oxy complex to form an Fe2+ cation intermediate. Deprotonation of the hydroxyl is demanding for both mechanisms and is assisted by Lys134 and Tyr101. | The tertiary structure also creates a metal binding site of Histidines to keep the iron molecule in place. There have been two mechanisms proposed for Lsd's. In one mechanism, the hydroxystillbenoid is activated via the enzyme-catalyzed deprotonation of the 4-hydroxy group, which then allows electron delocalization toward an Fe3+. In the other mechanism, π electron density from the double bond is redistributed to the iron-oxy complex to form an Fe2+ cation intermediate. Deprotonation of the hydroxyl is demanding for both mechanisms and is assisted by Lys134 and Tyr101. | ||
| - | + | ||
The hydrophobic/hydrophilic view of the ligand in the protein shows that both hydrophilic and hydrophobic residues are important to the ligand in the binding site. The first photo below shows the spacefill look at the protein, and it can be seen that the binding pocket is almost invisible and hard to reach. The second photo shows the hydrophobic/hydrophilic binding pocket up close, showing that the hydrophobic portion of the protein is interacting with the hydrophobic portion of the ligand, and the hydrophilic portion is interacting with the hydrophilic areas of the ligand. The red shows hydrophobic properties, and the green shows hydrophilic properties. In the first photo you can see on chain B the active site is mostly hydrophilic with one red hydrophobic area at the entrance. The ligand is in the middle of the protein and is not very visible from the outside of the protein. | The hydrophobic/hydrophilic view of the ligand in the protein shows that both hydrophilic and hydrophobic residues are important to the ligand in the binding site. The first photo below shows the spacefill look at the protein, and it can be seen that the binding pocket is almost invisible and hard to reach. The second photo shows the hydrophobic/hydrophilic binding pocket up close, showing that the hydrophobic portion of the protein is interacting with the hydrophobic portion of the ligand, and the hydrophilic portion is interacting with the hydrophilic areas of the ligand. The red shows hydrophobic properties, and the green shows hydrophilic properties. In the first photo you can see on chain B the active site is mostly hydrophilic with one red hydrophobic area at the entrance. The ligand is in the middle of the protein and is not very visible from the outside of the protein. | ||
| - | + | ||
| Line 39: | Line 39: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | <ref> 31292192 </ref> | + | <ref> PMID: 31292192 </ref> |
Revision as of 20:44, 8 December 2019
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
Overview
| |||||||||||