We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1558
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
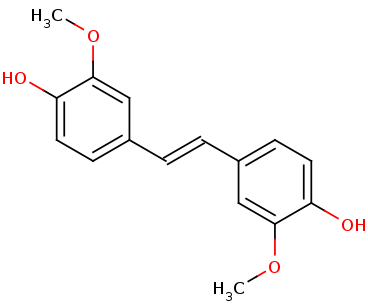
To examine LsdA’s substrate specificity, we heterologously produce the dimeric enzyme with the help of chaperones. When tested on several substituted stilbenes, LsdA exhibited the greatest specificity for lignostilbene. These experiments further indicate that the substrate’s | To examine LsdA’s substrate specificity, we heterologously produce the dimeric enzyme with the help of chaperones. When tested on several substituted stilbenes, LsdA exhibited the greatest specificity for lignostilbene. These experiments further indicate that the substrate’s | ||
4-hydroxy moiety is required for catalysis and that this moiety | 4-hydroxy moiety is required for catalysis and that this moiety | ||
| - | cannot be replaced with a methoxy group. This expands our | + | cannot be replaced with a methoxy group. This expands our mechanistic understanding of LsdA and related stilbene-cleaving dioxygenases. <ref>PMID 31292192</ref>. |
| - | mechanistic understanding of LsdA and related stilbene-cleaving dioxygenases. | + | |
== Structural highlights and structure-function relationships == | == Structural highlights and structure-function relationships == | ||
The <scene name='82/823082/Catalytic_triad/2'>Catalytic Triad</scene> of this protein is primarily made of the amino acids that are the main factor in catalysis. The 3 amino acids are Phe-59, Tyr101, and Lys-134. The <scene name='82/823082/Colored_secondary/1'>secondary and terteriary structure</scene> is a fold of LsdA of a seven-bladed -propeller, typical of the carotenoid cleavage oxygenates (CCO's), which usually catalyze the oxidative cleavage of a double bond in carotenoids. <ref>PMID 31292192</ref>. | The <scene name='82/823082/Catalytic_triad/2'>Catalytic Triad</scene> of this protein is primarily made of the amino acids that are the main factor in catalysis. The 3 amino acids are Phe-59, Tyr101, and Lys-134. The <scene name='82/823082/Colored_secondary/1'>secondary and terteriary structure</scene> is a fold of LsdA of a seven-bladed -propeller, typical of the carotenoid cleavage oxygenates (CCO's), which usually catalyze the oxidative cleavage of a double bond in carotenoids. <ref>PMID 31292192</ref>. | ||
| Line 18: | Line 17: | ||
The structures also consist of α-helices and ß-sheets. The <scene name='82/823082/Hydrogen_bonding/1'>active site</scene> occurs at the center of the propeller and contains an Fe2+. The <scene name='82/823082/Hydrophobicity_whole_protein/1'>hydrophobicity</scene> and <scene name='82/823082/Spacefill_whole_protein/1'>spacefill</scene> view of the ligand in the protein, which shows that both hydrophilic and hydrophobic residues are important to the ligand in the binding site. | The structures also consist of α-helices and ß-sheets. The <scene name='82/823082/Hydrogen_bonding/1'>active site</scene> occurs at the center of the propeller and contains an Fe2+. The <scene name='82/823082/Hydrophobicity_whole_protein/1'>hydrophobicity</scene> and <scene name='82/823082/Spacefill_whole_protein/1'>spacefill</scene> view of the ligand in the protein, which shows that both hydrophilic and hydrophobic residues are important to the ligand in the binding site. | ||
| - | The space fill view allows us to see the different binding sites for this protein, the binding site is allosteric. The binding site for this protein has a very restrictive accessibility. The <scene name='82/823082/Ligand_closeup/2'>ligand</scene> for this protein is called NSL. | + | The space fill view allows us to see the different binding sites for this protein, the binding site is allosteric. The binding site for this protein has a very restrictive accessibility. The <scene name='82/823082/Ligand_closeup/2'>ligand</scene> for this protein is called NSL. |
== Energy Transformation == | == Energy Transformation == | ||
Phenylazophenol inhibits the LsdA-catalyzed cleavage of lignostilbene in a reversible, mixed fashion. The substrate specificity studies of LsdA are consistent with | Phenylazophenol inhibits the LsdA-catalyzed cleavage of lignostilbene in a reversible, mixed fashion. The substrate specificity studies of LsdA are consistent with | ||
Current revision
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
Lignostilbene-α,β-dioxygenase A (LsdA) Catalyzation
| |||||||||||
References
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428