We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Riboswitch
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
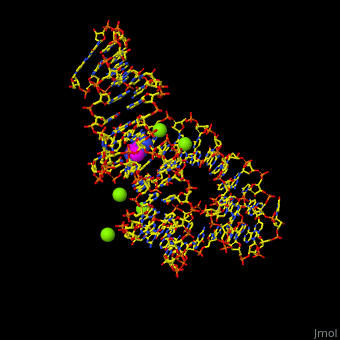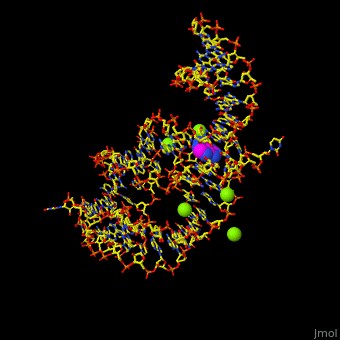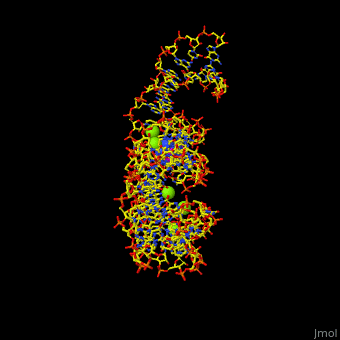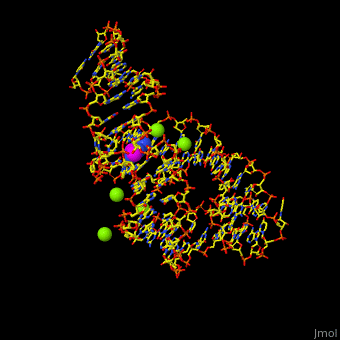
<StructureSection load='' size='350' side='right' scene='47/479252/Cv/1' caption='Adenine riboswitch complex with adenine and Mg+2 ions, [[1y26]]'> | <StructureSection load='' size='350' side='right' scene='47/479252/Cv/1' caption='Adenine riboswitch complex with adenine and Mg+2 ions, [[1y26]]'> | ||
| + | __TOC__ | ||
== Function == | == Function == | ||
Normally, a variety of proteins and protein cofactors control gene expression in an organism by binding to different sites on messenger RNA (mRNA). '''Riboswitches''' are genetic regulatory elements that are built directly into the [[RNA]]. They are a type of noncoding RNA that regulate gene expression in the absence of proteins by switching from one structural conformation (shape) to another in response to ligand binding. Most contain a single binding site that recognizes a specific ligand. The ability of a riboswitch to discriminate against molecules that are similar or closely related to its ligand is essential to prevent metabolic misregulation<ref name=scimag>Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. ''Science'', 319(5871), 1795-1797. doi:[http://dx.doi.org/10.1126/science.1152621 10.1126/science.1152621]</ref>. | Normally, a variety of proteins and protein cofactors control gene expression in an organism by binding to different sites on messenger RNA (mRNA). '''Riboswitches''' are genetic regulatory elements that are built directly into the [[RNA]]. They are a type of noncoding RNA that regulate gene expression in the absence of proteins by switching from one structural conformation (shape) to another in response to ligand binding. Most contain a single binding site that recognizes a specific ligand. The ability of a riboswitch to discriminate against molecules that are similar or closely related to its ligand is essential to prevent metabolic misregulation<ref name=scimag>Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. ''Science'', 319(5871), 1795-1797. doi:[http://dx.doi.org/10.1126/science.1152621 10.1126/science.1152621]</ref>. | ||
| Line 15: | Line 16: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of riboswitch== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Adenine riboswitch | ||
| - | |||
| - | **[[1y26]], [[4tzx]], [[4tzy]], [[4xnr]], [[5swd]], [[5swe]], [[5uza]] – VvAR + adenine – ''Vibrio vulnificus''<br /> | ||
| - | **[[5e54]] – VvAR <br /> | ||
| - | **[[3ivn]] – BsAR (mutant) – ''Bacillus subtilis''<br /> | ||
| - | **[[3la5]] - BsAR (mutant) + azacytosine<br /> | ||
| - | |||
| - | *Guanine riboswitch | ||
| - | |||
| - | **[[1y27]] – BsGR residues 185-252 + guanine<br /> | ||
| - | **[[2g9c]], [[3fo4]], [[3fo6]], [[3ges]], [[3gog]], [[3rkf]] - BsGR (mutant) + guanine derivative<br /> | ||
| - | **[[3got]] - BsGR (mutant) + adenine derivative<br /> | ||
| - | **[[2xo1]] - BsGR aptamer domain + adenine derivative<br /> | ||
| - | **[[3g4m]], [[3ger]] - BsGR + guanine derivative<br /> | ||
| - | **[[2xnz]] - BsGR aptamer domain + guanine derivative<br /> | ||
| - | **[[2xo0]] - BsGR aptamer domain + triazine derivative<br /> | ||
| - | **[[5c7u]], [[5c7w]] - BsGR + hypoxanthine<br /> | ||
| - | **[[2ees]], [[2eet]], [[2eeu]], [[2eev]], [[2eew]] - BsGR (mutant) + hypoxanthine<br /> | ||
| - | **[[3gao]] - BsGR + xanthine<br /> | ||
| - | **[[2xnw]] – BsGR + Mn<br /> | ||
| - | **[[4fe5]], [[4fej]], [[4fel]], [[4fen]], [[4feo]], [[4fep]] – GR + hypoxanthine – synthetic<br /> | ||
| - | |||
| - | *Guanidine riboswitch | ||
| - | |||
| - | **[[5nwq]] – TfGR + guanidine – ''Thermobifida fusca''<br /> | ||
| - | **[[5nzd]] – TfGR + CDP derivative<br /> | ||
| - | |||
| - | *Thiamine pyrophosphate riboswitch | ||
| - | |||
| - | **[[2gdi]] – TPPR + TPP – synthetic<br /> | ||
| - | **[[2hoj]], [[2hok]], [[2hol]] – EcTPPR + TPP + metal ion – ''Escherichia coli''<br /> | ||
| - | **[[2hom]] – EcTPPR + TMP<br /> | ||
| - | **[[2hoo]], [[2hop]] – EcTPPR + TPP analog<br /> | ||
| - | **[[4nya]], [[4nyb]], [[4nyc]] – EcTPPR + pyrimidine derivative<br /> | ||
| - | **[[4nyd]] – EcTPPR + hypoxanthine <br /> | ||
| - | **[[4nyg]] – EcTPPR + thyamine <br /> | ||
| - | **[[3d2g]], [[3d2v]], [[3d2x]] - TPPR + TPP analog – ''Arabidopsis thaliana'' | ||
| - | |||
| - | *S-adenosylmethionine riboswitch | ||
| - | |||
| - | **[[2gis]], [[4aob]], [[4b5r]] – TtSAMR + SAM – ''Thermoanaerobacter tengcongensis''<br /> | ||
| - | **[[3iqr]], [[2ydh]], [[2ygh]] - TtSAMR (mutant) + SAM<br /> | ||
| - | **[[3iqp]] - TtSAMR<br /> | ||
| - | **[[5fjc]], [[5fk1]], [[5fk2]], [[5fk3]], [[5fk4]], [[5fk5]], [[5fk6]], [[5fkd]], [[5fke]], [[5fkf]], [[5fkg]], [[5fkh]] - TtSAMR + SAM + GTP<br /> | ||
| - | **[[2qwy]], [[3e5c]], [[3e5e]], [[3e5f]], [[3iqn]] - SAMR + SAM – synthetic<br /> | ||
| - | **[[4kqy]] – BsSAMR + SAM<br /> | ||
| - | **[[4oqu]] - SAMR + SAM - synthetic<br /> | ||
| - | **[[6fz0]] - SAMR + SAM - ''Candidatus pelagibacter''<br /> | ||
| - | **[[6c27]] - SAMR + GTP – ''Enterococcus faecalis''<br /> | ||
| - | |||
| - | *S-adenosylhomocysteine riboswitch | ||
| - | |||
| - | **[[3npn]], [[3npq]] – SAHR + SAH – ''Ralstonia solanacearum''<br /> | ||
| - | **[[6hag]] - SAMR + SAH – proteobacteria - NMR<br /> | ||
| - | |||
| - | *Lysine riboswitch | ||
| - | |||
| - | **[[3dis]] – TmKR – ''Thermotoga maritima''<br /> | ||
| - | **[[3d0u]] – TmKR ligand-binding domain + Lysine <br /> | ||
| - | **[[3dil]], [[3dim]], [[3dio]], [[3dix]], [[3diy]], [[3diz]], [[3dj2]] – TmKR + Lys <br /> | ||
| - | **[[3dj0]], [[3dir]] – TmKR + Lys derivative<br /> | ||
| - | **[[3dig]] – TmKR + Cys derivative<br /> | ||
| - | **[[3diq]] – TmKR + homoArg <br /> | ||
| - | **[[4erj]] – TmKR + aminocaproic acid<br /> | ||
| - | **[[4erl]] – TmKR + Lys + Gly<br /> | ||
| - | **[[3d0x]] - KR - synthetic<br /> | ||
| - | |||
| - | *Glutamine riboswitch | ||
| - | |||
| - | **[[5ddp]] – QR + glutamine – human<br /> | ||
| - | **[[5ddq]], [[5ddr]] – SeQR + glutamine – ''Synechococcus elongatus''<br /> | ||
| - | **[[5ddo]] – SeQR + U1 small nuclear ribonucleoprotein<br /> | ||
| - | |||
| - | *FMN riboswitch | ||
| - | |||
| - | **[[3f2q]], [[3f2t]], [[3f2w]], [[3f2x]], [[3f2y]], [[3f30]], [[3f4e]] – FnFMNR + FMN – ''Fusobacterium nucleatum''<br /> | ||
| - | **[[2yie]] - FnFMNR aptamer domain + FMN<br /> | ||
| - | **[[3f4g]], [[3f4h]] – FnFMNR + flavin derivative<br /> | ||
| - | **[[2yif]] - FnFMNR + GTP<br /> | ||
| - | **[[6dn1]], [[6dn2]], [[6dn3]] - FnFMNR + RNA<br /> | ||
| - | **[[5c45]], [[5kx9]], [[6bfb]] - FnFMNR + inhibitor<br /> | ||
| - | |||
| - | *Pre-queosine riboswitch | ||
| - | |||
| - | **[[3fu2]], [[3k1v]] – BsQ1R + queosine<br /> | ||
| - | **[[3gca]] – TtQ0R + queosine<br /> | ||
| - | **[[3q50]], [[3q51]] – TtQ1R aptamer domain + queosine<br /> | ||
| - | **[[4jf2]], [[5d5l]] – Q1R + GTP + deaza-aminomethyl guanine – ''Lactobacillus rhamnosus''<br /> | ||
| - | **[[4rzd]] – Q1R + GTP + deaza-aminomethyl guanine – ''Faecalibacterium prausnitzii''<br /> | ||
| - | |||
| - | *C-di-GMP riboswitch | ||
| - | |||
| - | **[[3irw]], [[3mxh]] – VcGMPR + C-di-GMP + GTP + U1 small nuclear ribonucleoprotein – ''Vibrio cholerae''<br /> | ||
| - | **[[3iwn]], [[4yb1]] – VcGMPR + C-di-GMP + U1 small nuclear ribonucleoprotein<br /> | ||
| - | **[[3mum]], [[3mur]], [[3mut]] - VcGMPR (mutant) + C-di-GMP + U1 small nuclear ribonucleoprotein<br /> | ||
| - | **[[3muv]] - VcGMPR (mutant) + C-di-AMP + U1 small nuclear ribonucleoprotein<br /> | ||
| - | **[[3q3z]] - GMPR + C-di-GMP – ''Clostridium acetobutylicum''<br /> | ||
| - | **[[4qk8]], [[4qka]] – GMPR + GTP – ''Thermoanaerobacter pseudethanolicus''<br /> | ||
| - | **[[4qk9]] – GMPR + guanosine derivative – ''Thermovirga lienii''<br /> | ||
| - | **[[4yaz]], [[4yb0]] – GMPR + C-di-GMP - ''Geobacter''<br /> | ||
| - | |||
| - | *C-di-AMP riboswitch | ||
| - | |||
| - | **[[4w90]], [[4w92]] – VBsAMPR + C-di-AMP + U1 small nuclear ribonucleoprotein<br /> | ||
| - | **[[3muv]] – AMPR + C-di-AMP + U1 small nuclear ribonucleoprotein - synthetic<br /> | ||
| - | **[[4qk8]] – AMPR + C-di-AMP + GTP - ''Thermoanaerobacter pseudethanolicus''<br /> | ||
| - | **[[4qk8]], [[4qka]] – AMPR + C-di-AMP + GTP - ''Thermoanaerobacter pseudethanolicus''<br /> | ||
| - | **[[4qlm]], [[4qln]] – AMPR + C-di-AMP - ''Thermoanaerobacter tengcongensis''<br /> | ||
| - | **[[4qk9]] – AMPR + C-di-AMP + GTP derivative - ''Thermovirga lienii''<br /> | ||
| - | |||
| - | *Glycine riboswitch | ||
| - | |||
| - | **[[3owi]], [[3oww]], [[3owz]] – VcGlyR + glycine<br /> | ||
| - | **[[3ox0]], [[3oxe]], [[3oxj]], [[3oxm]] – VcGlyR + GDP + cytidine cyclic phosphate<br /> | ||
| - | **[[3oxb]], [[3oxd]] – VcGlyR (mutant) + GDP + cytidine cyclic phosphate<br /> | ||
| - | **[[3p49]] – FnGlyR + U1 small nuclear ribonucleoprotein + glycine<br /> | ||
| - | |||
| - | *M-Box riboswitch | ||
| - | |||
| - | **[[3pdr]] – BsMBR + Mn<br /> | ||
| - | **[[2qbz]] – BsMBR + Mg<br /> | ||
| - | |||
| - | *T-Box riboswitch | ||
| - | |||
| - | **[[4lck]] – sTBR + tRNA-Gly + ribosomal protein YBXF – synthetic<br /> | ||
| - | **[[4mgn]] – sTBR + tRNA-Gly <br /> | ||
| - | |||
| - | *Tetrahydrofolate riboswitch | ||
| - | |||
| - | **[[3suh]], [[3sux]] – EsTHFR + THF derivative – ''Eubacterium siraeum''<br /> | ||
| - | **[[3suy]] - EsTHFR + cytidine cyclic phosphate<br /> | ||
| - | **[[4lvv]], [[4lvx]], [[4lvw]], [[4lvy]], [[4lvz]], [[4lw0]] – THFR + purine derivative – synthetic<br /> | ||
| - | **[[3sd3]] – THFR (mutant) + pteridine derivative – synthetic<br /> | ||
| - | |||
| - | *Fluoride riboswitch | ||
| - | |||
| - | **[[3vrs]] – TpR + Mn + K + F – ''Thermotoga petrophila''<br /> | ||
| - | **[[4en5]] – TpR + Mg + Tl + F <br /> | ||
| - | **[[4enc]] – TpR + Mg + K + F <br /> | ||
| - | **[[4ena]] – TpR + Mg + Cs + GTP + F <br /> | ||
| - | **[[4enb]] – TpR + Mg + Ir + K + GTP + F <br /> | ||
| - | **[[5kh8]] – R - synthetic - NMR<br /> | ||
| - | |||
| - | *Cobalamine riboswitch | ||
| - | |||
| - | **[[4frg]], [[4frn]], [[4gma]], [[4gxy]] – CoR + aptamer domain – ''Marine metagenome''<br /> | ||
| - | |||
| - | *yybp-ykoy riboswitch | ||
| - | |||
| - | **[[4y1i]] – LlYYR + Mn – ''Lactobacillus lactis''<br /> | ||
| - | **[[4y1j]] – LlYYR (mutant) + Mn<br /> | ||
| - | **[[4y1m]] – EcYYR <br /> | ||
| - | |||
| - | *ykkc riboswitch | ||
| - | |||
| - | **[[5u3g]] – TpR + gusanidine – ''Dikeya dadantii''<br /> | ||
| - | |||
| - | *PRPP riboswitch | ||
| - | |||
| - | **[[6ck5]] – TmPRR + PRPP – ''Thermoanaerobacter mathranii''<br /> | ||
| - | **[[6ck4]] – TmPRR + PPGPP<br /> | ||
| - | **[[6dnr]] – SlPRR + GTP + cytidine phosphate – ''Syntrophothermus lipocalidus''<br /> | ||
| - | **[[6dlq]], [[6dls]], [[6dlt]] – SlPRR + GTP + cytidine phosphate + PRPP<br /> | ||
| - | **[[6dlr]] – SlPRR + cytidine phosphate + PRPP<br /> | ||
| - | |||
| - | * PPGPP riboswitch | ||
| - | |||
| - | **[[6dme]], [[6dmd]], [[6dmc]] – SaPPGPPR + GTP + cytidine phosphate + guanine tetraphosphate – ''Sulfobacillus acidophilus''<br /> | ||
| - | |||
| - | *PFI riboswitch | ||
| - | |||
| - | **[[4znp]] – TpR + AICAR – synthetic<br /> | ||
| - | |||
| - | *NICO riboswitch | ||
| - | |||
| - | **[[4rum]] – TpR + Co – synthetic<br /> | ||
| - | |||
| - | *ZMP riboswitch | ||
| - | |||
| - | **[[4xw7]], [[4xwf]] – TpR + AICAR – ''Actinomyces odontolyticus'' <br /> | ||
| - | |||
| - | *ZTP riboswitch | ||
| - | |||
| - | **[[5btp]] – TpR + AICAR – ''Fusobacterium ulcerans'' <br /> | ||
| - | |||
| - | *Guanidine riboswitch | ||
| - | **[[5t83]] – TpR + guanidine – ''Sulfobacillus acidophilus'' <br /> | ||
| - | **[[5ndh]], [[5ndi]], [[5nef]], [[5neo]], [[5nep]], [[5neq]], [[5nex]], [[5nom]] – TpR + guanidine derivative – ''Gloeobacter violaceus'' <br /> | ||
| - | **[[5vj9]], [[5vjb]] – TpR + guanidine – ''Pseudomonas aeruginosa'' <br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category: Topic Page]] | [[Category: Topic Page]] | ||
Revision as of 10:03, 31 December 2019
| |||||||||||
References
- ↑ Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. Science, 319(5871), 1795-1797. doi:10.1126/science.1152621
- ↑ Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004 Dec;11(12):1729-41. PMID:15610857 doi:S1074-5521(04)00343-6
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Karsten Theis, Joel L. Sussman