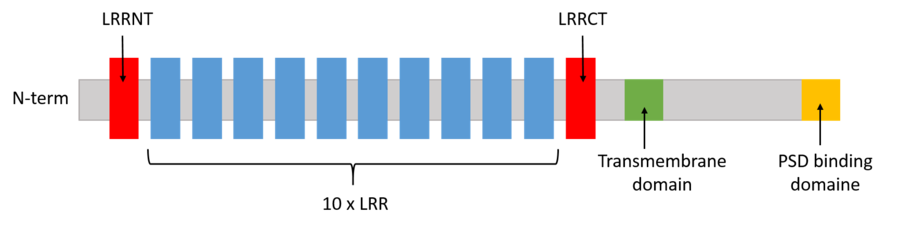Sandbox Reserved 1093
From Proteopedia
| Line 10: | Line 10: | ||
[[Image:LRRTM2 details.png|900px|left]] | [[Image:LRRTM2 details.png|900px|left]] | ||
| - | 1) <scene name='82/829346/Lrrtm2/ | + | 1) <scene name='82/829346/Lrrtm2/5'>N-term fixation domain</scene> |
The N-terminal signal peptide is long of 33 amino. The extracellular domain contains 399 amino acids organized in 2 cysteine-rich domains (<scene name='82/829346/Lrrnt/1'>LRRNT</scene> and <scene name='82/829346/Lrrct/1'>LRRCT</scene>) and <scene name='82/829346/Lrr/2'>10 leucine rich domains</scene> (LRR). Each LRR domain is composed of 21 amino acids containing the conserved 11-aa sequence, LxxLxLxxN/ CxL, where x is any amino acid, and <scene name='82/829346/Leucine/1'>leucine</scene> and asparagine can be replaced with other hydrophobic residues. The leucine rich repeat domain forms a convex structure stabilized by a <scene name='82/829346/Phe/1'>Phe</scene> spine. The concave surface is composed of a continuous <scene name='82/829346/Beta_sheets/1'>β-sheet</scene>, which provides an effective ligand-binding site, whereas the convex surface consists of <scene name='82/829346/Alpha_helix/1'>α-helices</scene>, which affect the curvature of the LRR domain. | The N-terminal signal peptide is long of 33 amino. The extracellular domain contains 399 amino acids organized in 2 cysteine-rich domains (<scene name='82/829346/Lrrnt/1'>LRRNT</scene> and <scene name='82/829346/Lrrct/1'>LRRCT</scene>) and <scene name='82/829346/Lrr/2'>10 leucine rich domains</scene> (LRR). Each LRR domain is composed of 21 amino acids containing the conserved 11-aa sequence, LxxLxLxxN/ CxL, where x is any amino acid, and <scene name='82/829346/Leucine/1'>leucine</scene> and asparagine can be replaced with other hydrophobic residues. The leucine rich repeat domain forms a convex structure stabilized by a <scene name='82/829346/Phe/1'>Phe</scene> spine. The concave surface is composed of a continuous <scene name='82/829346/Beta_sheets/1'>β-sheet</scene>, which provides an effective ligand-binding site, whereas the convex surface consists of <scene name='82/829346/Alpha_helix/1'>α-helices</scene>, which affect the curvature of the LRR domain. | ||
| Line 33: | Line 33: | ||
'''LRRTMs Family
''' | '''LRRTMs Family
''' | ||
| - | All four members of the human LRRTM family are highly similar in their LRR domains with >55% sequence identity. But only LRRTM1 and LRRTM2 have been extensively studied in the context of the interaction with Nrxn. This is due to the fact that some critical residues for binding with Nrxn1β have been replaced in LRRTM3 and LRRTM4, such as <scene name='82/829346/Glu_348/1'>Glu348</scene>, <scene name='82/829346/Asp_352/1'>Asp352</scene>, and <scene name='82/829346/Phe_357/2'>Phe357</scene>. | + | All four members of the human LRRTM family are highly similar in their LRR domains with >55% sequence identity. But only LRRTM1 and LRRTM2 have been extensively studied in the context of the interaction with Nrxn. This is due to the fact that some critical residues for binding with Nrxn1β [http://proteopedia.org/wiki/index.php/Neurexin] have been replaced in LRRTM3 and LRRTM4, such as <scene name='82/829346/Glu_348/1'>Glu348</scene>, <scene name='82/829346/Asp_352/1'>Asp352</scene>, and <scene name='82/829346/Phe_357/2'>Phe357</scene>. |
The replacement of Glu348 by Val in LRRTM3 is likely to prevent of the interaction between Ca2+ and Nrxn1β. It is possible that other specific residue(s) of LRRTM3/4 may also prevent the binding. | The replacement of Glu348 by Val in LRRTM3 is likely to prevent of the interaction between Ca2+ and Nrxn1β. It is possible that other specific residue(s) of LRRTM3/4 may also prevent the binding. | ||
'''Ligands''' | '''Ligands''' | ||
| - | The structure of the complex Nrxn1β–LRRTM2[http://proteopedia.org/wiki/index.php/5z8y]is being determined by co-crystallisation. A mutation from His 355 to Ala 355 without affecting the complex structure is necessary to maintain the stability of the crystal. | + | The structure of the complex <scene name='82/829346/Complex/1'>Nrxn1β–LRRTM2</scene>[http://proteopedia.org/wiki/index.php/5z8y]is being determined by co-crystallisation. A mutation from His 355 to Ala 355 without affecting the complex structure is necessary to maintain the stability of the crystal. |
| Line 47: | Line 47: | ||
Cbln1–GluD2 | Cbln1–GluD2 | ||
| + | |||
| + | '''Neurexins''' | ||
| + | |||
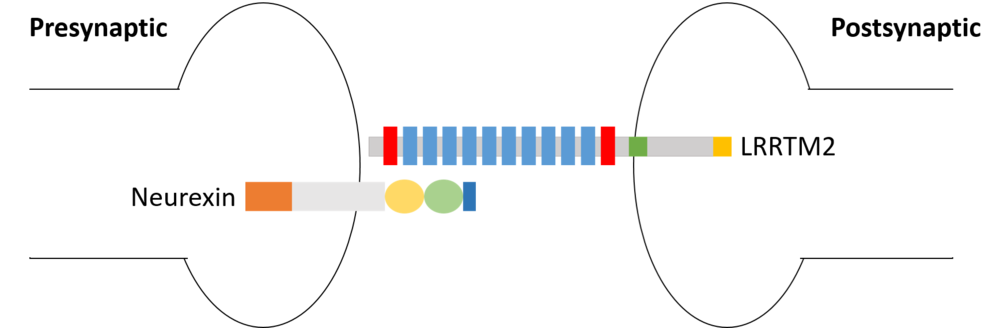
| + | Neurexins (Nrxns) [http://proteopedia.org/wiki/index.php/Neurexin]is a family of presynaptic organizer which interact with several postsynaptic organizers such as LRRTM2. | ||
| + | There are three Neurexin genes in vertebrates, each corresponding to a different promoter type. Neurexins are characterized by their laminin-neurexin-sex hormone (LNS) domains. ︎ <scene name='82/829346/Neurexin_1_alpha/1'>α-neurexins</scene> have six whereas ︎<scene name='82/829346/Beta-neurexin_1/2'>β-neurexins</scene> have a single LNS domain. The α-helical conformation causes severe steric hindrance with the bound LRRTM2, whereas the β-stranded conformation causes no obvious steric hindrance. | ||
== Disease == | == Disease == | ||
| Line 52: | Line 57: | ||
A large number of researches shows that LRRTM2 is related to bipolar disorder. | A large number of researches shows that LRRTM2 is related to bipolar disorder. | ||
A deletion (240 kb) at 5q31 chromosomal region containing LRRTM2 and CTNNA1 has been shown to be related to intellectual disability and developmental delay. | A deletion (240 kb) at 5q31 chromosomal region containing LRRTM2 and CTNNA1 has been shown to be related to intellectual disability and developmental delay. | ||
| - | |||
| - | |||
| - | '''Neurexins''' | ||
| - | |||
| - | Neurexins (Nrxns) is a presynaptic organizer family which interact with several postsynaptic organizers. | ||
| - | There are three Neurexin genes in vertebrates, each corresponds to a different promoter. Neurexins are characterized by their laminin-neurexin-sex hormone (LNS) domains. ︎ α-neurexins have six whereas ︎β-neurexins have a single LNS domain. The α-helical conformation causes severe steric hindrance with the bound LRRTM2, whereas the β-stranded conformation causes no obvious steric hindrance. | ||
| - | |||
== References == | == References == | ||
Revision as of 12:23, 15 January 2020
is a transmembrane protein that can be found in human neurons. It functions as postsynaptic organizers that induce excitatory synapses. LRRTM2 is prominently expressed in deep layers, rather than superficial layers, of the cerebral cortex. LRRTM2 specifically localizes in excitatory synapses, and not in inhibitory synapses. In addition, LRRTMs interact with neurexins[1]to bridge the synaptic cleft.
Contents |
Structural highlights
| |||||||||||
Function
Related Structures
LRRTMs Family
All four members of the human LRRTM family are highly similar in their LRR domains with >55% sequence identity. But only LRRTM1 and LRRTM2 have been extensively studied in the context of the interaction with Nrxn. This is due to the fact that some critical residues for binding with Nrxn1β [2] have been replaced in LRRTM3 and LRRTM4, such as , , and . The replacement of Glu348 by Val in LRRTM3 is likely to prevent of the interaction between Ca2+ and Nrxn1β. It is possible that other specific residue(s) of LRRTM3/4 may also prevent the binding.
Ligands
The structure of the complex [3]is being determined by co-crystallisation. A mutation from His 355 to Ala 355 without affecting the complex structure is necessary to maintain the stability of the crystal.
Other synaptic organisers
�
Neuroligins (NLs)
LRRTM2 bind to Neurexins 1, 2 and 3 ︎and ︎a variant region at splice site 4 in the LNS. As the variant region lacking a 30 amino acid insert (-S4), LRRTM2 cannot induce presynaptic differentiation in neurons. On the contrary, Neuroligin1 binds to Neurexins 1, 2, and 3 ︎ but not ︎to variants, has a higher affinity with Neurexin 1 (-S4) than with Neurexin 1 (+S4)
Cbln1–GluD2
Neurexins
Neurexins (Nrxns) [4]is a family of presynaptic organizer which interact with several postsynaptic organizers such as LRRTM2. There are three Neurexin genes in vertebrates, each corresponding to a different promoter type. Neurexins are characterized by their laminin-neurexin-sex hormone (LNS) domains. ︎ have six whereas ︎ have a single LNS domain. The α-helical conformation causes severe steric hindrance with the bound LRRTM2, whereas the β-stranded conformation causes no obvious steric hindrance.
Disease
A large number of researches shows that LRRTM2 is related to bipolar disorder. A deletion (240 kb) at 5q31 chromosomal region containing LRRTM2 and CTNNA1 has been shown to be related to intellectual disability and developmental delay.
References
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6160412/ https://pdf.sciencedirectassets.com/271175/1-s2.0-S0168010217X00047/1-s2.0-S0168010216302176/main.pdf?X-Amz-Security-Token=IQoJb3JpZ2luX2VjEIn%2F%2F%2F%2F%2F%2F%2F%2F%2F%2FwEaCXVzLWVhc3QtMSJIMEYCIQD9y3Gq4IpA3WUL5u%2Fkg2WA1Xkc%2FecGOKwBvrh87jA43QIhALaUG9EO6UsgdfDX4BdAFQdHgdcRASV4GY5gcp4MpAU1KrQDCBEQAhoMMDU5MDAzNTQ2ODY1IgxX8k4XAFbLEwxUIxEqkQMGYQSzdSg4KJygQuQhcirZ5z1dcUiJllkhebembjnSpLm2HgwQyXo8kS7OyOG4LrZK%2FpuVLgcwKJPzhlzfC8hvL4XkbdOHINPOAHjqrQAZfDUTyerG37EygqlyBH3ozWLj6bBRzb4qjtTKHiJXIVViFUwE4kLnUx%2BG1P9nlMZKiKwjTTZANO6qdo02b0eBH5wtGZXkTThixMrkac5AkC%2F6lv55c6GQkaGJ7QFUTzuMDhw1jnjgjh3SYEvL3zSXYMMmK9cdAvX47pXUxrx2upPm%2B1b6tXK9t%2BtcMhmGekMeq%2BQ4vgAGco9W47wKMZckdEtWsBwLD0ouczegSiUsY2j7%2Bkbrq3doyu8IVfj2trxYgsDhsot0o9LFT6vU4OcBDau43lRiWt28NL9taG2HVIr6S0JpdQrG5GnhP%2B9JBw0NzpNienHWZklAjtH5Yf2UdelMJoVxQVhA5Wt%2BxJdRYEa96OmlsXN%2FPrTFepkkdCM8oRnTDPYIsrzdNE7ztyE%2BKdycj4X5AmvBRpmLhlhj8JCajzDXhObwBTrqAbLxyZNQ%2F%2BztbwIwb1i%2FYTwtf4elBbvP75%2F%2BPavRxseS9SYjHMist7P2A3ic9sXaaRIVHQ%2BiNot7dRJXTHEmnQm%2Fpv7wJS%2BL3FrBNKhq2dtEs3wZwYiSkGRlQcFqq9B%2F6z%2FEpYEhZyY%2BXvVFlPy3UBFWSae%2B7sI6WH7Ioqesc5tp20wMy%2B86dD5OmNOpvKjmiRLEEzQU3yDhDHJbx3MEUlMTaugj2oDUpQwOhwwHGH5RWTUcyZQY2%2FZXTfvrmb4CbE9z%2BStJMmKQNGFFSfwmHo5tGI5vdQhOhljI0Jbg6Uarx7PMrUlHTCMy%2FA%3D%3D&X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Date=20200111T090940Z&X-Amz-SignedHeaders=host&X-Amz-Expires=300&X-Amz-Credential=ASIAQ3PHCVTYRBGSTTXK%2F20200111%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Signature=f3a0fb03947cf2be7f5c0d563d999a97cbc798dfcf6a9362970a2b4f22429f84&hash=733b1d2a78272d2393866d8fc6492b461308206ea979170d41dd5fa61f0ff15f&host=68042c943591013ac2b2430a89b270f6af2c76d8dfd086a07176afe7c76c2c61&pii=S0168010216302176&tid=spdf-83ad8d7e-b6c3-48f3-ac67-86f9a916e1d5&sid=45412b395b1d0047476b49a0dd1ef07e3ef6gxrqb&type=client https://www.sciencedirect.com/science/article/pii/S2211124715015375 https://www.sciencedirect.com/science/article/abs/pii/S0959438810001364#! https://www.karger.com/Article/FullText/341252 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6160412/ https://www.uniprot.org/uniprot/O43300 https://books.google.fr/books?id=hLS9BAAAQBAJ&pg=PA325&lpg=PA325&dq=LRRTM2+cytoplasmic+domain&source=bl&ots=5qlA_emVJs&sig=ACfU3U3tO8IN9lWW0rq3DDJbolvaJAo6Tw&hl=fr&sa=X&ved=2ahUKEwij9K_eofLmAhVCqxoKHZzPD8oQ6AEwAnoECAsQAQ#v=onepage&q=LRRTM2%20cytoplasmic%20domain&f=false - Cell Adhesion Molecules: Implications in Neurological Diseases publié par Vladimir Berezin, Peter S. Walmod https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3887770/ https://onlinelibrary.wiley.com/doi/pdf/10.1111/jnc.13159 https://www.rcsb.org/structure/5Z8X