We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1102
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
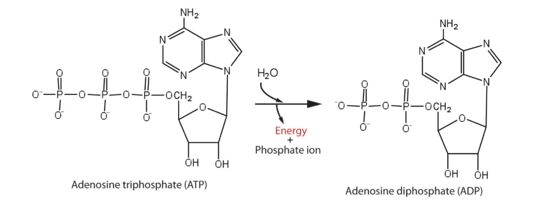
Protein from Zika is composed of a protease domain at its [https://en.wikipedia.org/wiki/N-terminus N terminus] and a helicase domain at its [https://en.wikipedia.org/wiki/C-terminus C terminus]. The multifunctional helicase belongs to the superfamily 2 (SF2) helicase family. It is this helicase domain that has 5'-triphosphatase activity and that performs the critical and indispensable function of unwinding double-stranded RNA during Zika genome replication. So it is an essential enzyme involved in the cycles of [https://en.wikipedia.org/wiki/ATP_hydrolysis ATP hydrolysis] and behaves as a motor protein in the RNA unwinding. Helicase performs ATP hydrolysis at the 5′ end of RNA to generate energy. The phosphorus atom of the γ-phosphate group of the ATP molecule is attacked by a water molecule; then, the γ-phosphate group is released with ADP. With the use of the energy derived from ATP hydrolysis, helicase translocates along nucleic acid strands and unwinds the double-stranded RNA genome. | Protein from Zika is composed of a protease domain at its [https://en.wikipedia.org/wiki/N-terminus N terminus] and a helicase domain at its [https://en.wikipedia.org/wiki/C-terminus C terminus]. The multifunctional helicase belongs to the superfamily 2 (SF2) helicase family. It is this helicase domain that has 5'-triphosphatase activity and that performs the critical and indispensable function of unwinding double-stranded RNA during Zika genome replication. So it is an essential enzyme involved in the cycles of [https://en.wikipedia.org/wiki/ATP_hydrolysis ATP hydrolysis] and behaves as a motor protein in the RNA unwinding. Helicase performs ATP hydrolysis at the 5′ end of RNA to generate energy. The phosphorus atom of the γ-phosphate group of the ATP molecule is attacked by a water molecule; then, the γ-phosphate group is released with ADP. With the use of the energy derived from ATP hydrolysis, helicase translocates along nucleic acid strands and unwinds the double-stranded RNA genome. | ||
[[Image:ATPhydrolysis.jpg | thumb | upright=3,5 | Energy from ATP hydrolysis]] | [[Image:ATPhydrolysis.jpg | thumb | upright=3,5 | Energy from ATP hydrolysis]] | ||
| - | This is | + | This step is among the necessary steps when it comes to viral RNA replication. It provides, then, conditions for the polymerization of RNA by an RNA-dependent RNA polymerase and the methylation of RNA by methyltransferase, which is essential for Zika replication. Moreover, it has been noticed that the viral replicative capacity and efficiency can be modified by the ATPase activity and therefore. Moreover, this enzyme can also change the host innate immune response. |
| - | ATP hydrolysis | + | ATP hydrolysis constitutes the most basic event of this helicase’s function, however, the other dynamic mechanisms remain largely unknown, impeding the further understanding of the function of Zika helicase. |
<ref> Hongliang Tian, Xiaoyun Ji, Xiaoyun Yang, Zhongxin Zhang, Zuokun Lu, Kailin Yang, Cheng Chen, Qi Zhao, Heng Chi, Zhongyu Mu, Wei Xie, Zefang Wang, Huiqiang Lou, Haitao Yang and Zihe Rao. Structural basis of Zika virus helicase in recognizing its substrates[J]. Protein&Cell, 2016, 7(8): 562-570. doi: 10.1007/s13238-016-0293-2 http://www.protein-cell.org/en/article/doi/10.1007/s13238-016-0293-2 </ref> | <ref> Hongliang Tian, Xiaoyun Ji, Xiaoyun Yang, Zhongxin Zhang, Zuokun Lu, Kailin Yang, Cheng Chen, Qi Zhao, Heng Chi, Zhongyu Mu, Wei Xie, Zefang Wang, Huiqiang Lou, Haitao Yang and Zihe Rao. Structural basis of Zika virus helicase in recognizing its substrates[J]. Protein&Cell, 2016, 7(8): 562-570. doi: 10.1007/s13238-016-0293-2 http://www.protein-cell.org/en/article/doi/10.1007/s13238-016-0293-2 </ref> | ||
Revision as of 16:20, 16 January 2020
| This Sandbox is Reserved from 25/11/2019, through 30/9/2020 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1091 through Sandbox Reserved 1115. |
To get started:
More help: Help:Editing |
5y6n- Zika virus helicase in complex with ADP
5y6n is a 1 chain protein structure. It’s the only helicase that belongs to zika virus. Zika helicase plays an important role in the pathogenocity of this virus.
| |||||||||||