Sandbox Reserved 1598
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
[[User: Grace A. Bassler/Sandbox 1]] | [[User: Grace A. Bassler/Sandbox 1]] | ||
{{Sandbox_Reserved_CH462_Biochemistry_II}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_CH462_Biochemistry_II}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | = | + | =bd Oxidase= |
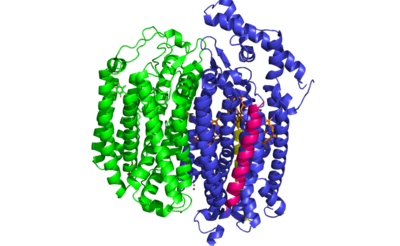
| - | <StructureSection load=' | + | <StructureSection load='6RX4' size='350' frame='true' side='right' caption='bd Oxidase: 6RX4' scene=’’> |
| - | + | <scene name='83/837228/Bd_oxidase/1'>bd Oxidase</scene> | |
| - | + | ||
| - | == | + | == Introduction == |
| - | + | [https://en.wikipedia.org/wiki/Escherichia_coli ''E. coli''] | |
| + | |||
| + | In-text citation<ref name="Ransey">PMID:28504306</ref>. | ||
| + | Second in-text citation<ref name="Safarian">PMID: 27126043</ref>. | ||
| + | |||
| + | [[Image:cyd s.png|400 px|right|thumb|Figure 1: bd Oxidase Subunits.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Structure == | ||
| + | |||
| + | === Subunits === | ||
| + | |||
| + | ''E. coli'' bd oxidase is made up of four individual subunits. The two major subunits, CydA and CydB, are each composed of one peripheral helix and two bundles of four transmembrane helices. The <scene name='83/837228/Cyda/3'>CydA subunit</scene> plays the most important role in the oxygen reduction reaction as it contains the Q-loop as well as all three heme groups. The <scene name='83/837228/Cydb/1'>CydB subunit</scene> harbors the ubiquinone molecule which provides structural support to the subunit that mimics the three hemes found in CydA. The remaining two subunits, CydS and CydX, are both single helix structures that assist in the oxygen reduction reaction. Unique to ''E. coli'', the <scene name='83/837228/Cyds/1'>CydS subunit</scene> binds to CydA to block oxygen from directly binding to heme b595. The <scene name='83/837228/Cydx/1'>CydX subunit</scene> promotes the assembly and stability of the oxidase complex. | ||
| + | |||
| + | |||
| + | insert citations & external links!** | ||
| + | |||
| + | |||
| + | ===Q Loop=== | ||
| + | |||
| + | <scene name='83/837228/Q_loop/1'>Q Loop</scene> | ||
| + | |||
| + | === Hemes === | ||
| + | |||
| + | |||
| + | == Function == | ||
== Disease == | == Disease == | ||
| Line 17: | Line 49: | ||
== Structural highlights == | == Structural highlights == | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
| + | <ref name="Ransey">PMID:28504306</ref>. | ||
| + | <ref name="Safarian">PMID: 27126043</ref>. | ||
<references/> | <references/> | ||
Revision as of 19:26, 30 March 2020
User: Grace A. Bassler/Sandbox 1
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
bd Oxidase
| |||||||||||
References
- ↑ 1.0 1.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ 2.0 2.1 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477