This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Transketolase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
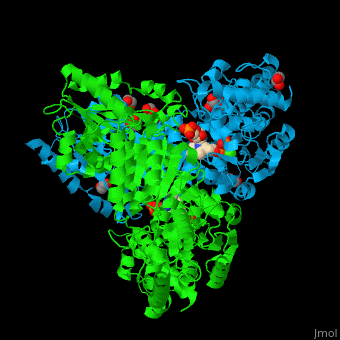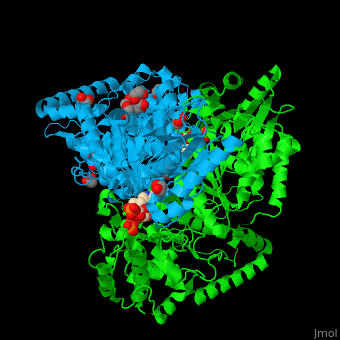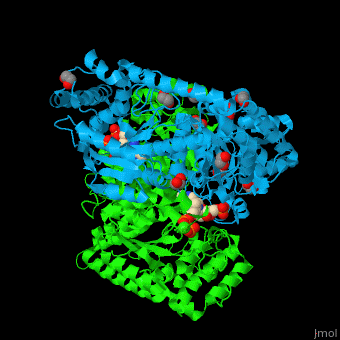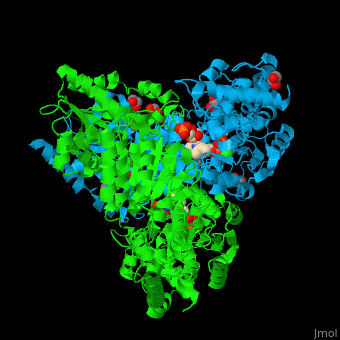
<StructureSection load='' size='350' side='right' caption='E. coli transketolase 1 dimer complex with xylulose-5-phosphate-thiamine diphosphate adduct, ethylene glycol and Ca+2 ion (green) (PDB code [[2r8o]]).' scene='46/466534/Cv/1'> | <StructureSection load='' size='350' side='right' caption='E. coli transketolase 1 dimer complex with xylulose-5-phosphate-thiamine diphosphate adduct, ethylene glycol and Ca+2 ion (green) (PDB code [[2r8o]]).' scene='46/466534/Cv/1'> | ||
== Function == | == Function == | ||
| - | '''Transketolase''' (TKT) catalyzes two opposite reactions. The synthesis of sedoheptulose-7-P in the pentose phosphate pathway using thiamine diphosphate (TPP) as co-factor; and the conversion of sedoheptulose-7-P and glyceraldehyde-3-P to aldose and ketose in the Calvin cycle<ref>PMID:8369276</ref>. | + | '''Transketolase''' (TKT) catalyzes two opposite reactions. The synthesis of sedoheptulose-7-P in the pentose phosphate pathway using thiamine diphosphate (TPP) as co-factor; and the conversion of sedoheptulose-7-P and glyceraldehyde-3-P to aldose and ketose in the Calvin cycle<ref>PMID:8369276</ref>. Split-gene TKT is found in the hyperthermophilic bacterium ''Carboxydothermus hydrogenoformans'' and is reconstituted from 2 separate polypeptide chains<ref>PMID:33193259</ref>. |
== Structural highlights == | == Structural highlights == | ||
| Line 48: | Line 48: | ||
**[[1qgd]] - EcTKT + TPP <br /> | **[[1qgd]] - EcTKT + TPP <br /> | ||
| + | **[[6tj8]] - EcTKT 1 + TPP analog <br /> | ||
**[[5hht]] - EcTKT (mutant) + TDP <br /> | **[[5hht]] - EcTKT (mutant) + TDP <br /> | ||
**[[1r9j]] - TKT + TPP – ''Leishmania mexicana''<br /> | **[[1r9j]] - TKT + TPP – ''Leishmania mexicana''<br /> | ||
**[[4c7x]] - LsTKT + TPP <br /> | **[[4c7x]] - LsTKT + TPP <br /> | ||
| - | **[[5hyv]], [[5x56]], [[5xuf]], [[5xvt]] - PsTKT + TPP <br /> | + | **[[5hyv]], [[5x56]], [[5xuf]], [[5xvt]], [[5xs6]] - PsTKT + TPP <br /> |
**[[5xsa]], [[5xsb]], [[5xsm]], [[5xt0]], [[5xt4]], [[5xtv]], [[5xtx]] - PsTKT + TPP intermediate<br /> | **[[5xsa]], [[5xsb]], [[5xsm]], [[5xt0]], [[5xt4]], [[5xtv]], [[5xtx]] - PsTKT + TPP intermediate<br /> | ||
| + | **[[5xqk]] - PsTKT + xylulose-5-P<br /> | ||
| + | **[[5xqa]] - PsTKT + ribose-5-P<br /> | ||
| + | **[[5xps]] - PsTKT + erythrose-4-P<br /> | ||
| + | **[[5xrv]] - PsTKT + fructose-6-P<br /> | ||
| + | **[[5i51]] - PsTKT (mutant) + fructose-6-P<br /> | ||
**[[5xtl]] - PsTKT + pyrimidine derivative <br /> | **[[5xtl]] - PsTKT + pyrimidine derivative <br /> | ||
**[[5nd5]] - CrTKT + TPP <br /> | **[[5nd5]] - CrTKT + TPP <br /> | ||
**[[3m49]] – BaTKT + TPP<br /> | **[[3m49]] – BaTKT + TPP<br /> | ||
| - | + | ||
*Transketolase ternary complexes | *Transketolase ternary complexes | ||
| Line 70: | Line 76: | ||
**[[2r5n]] – EcTKT 1 + TPP + ribose 5-P<br /> | **[[2r5n]] – EcTKT 1 + TPP + ribose 5-P<br /> | ||
**[[3upt]],[[5xqa]] - BtTKT + TPP + ribose 5-P<br /> | **[[3upt]],[[5xqa]] - BtTKT + TPP + ribose 5-P<br /> | ||
| - | **[[2r8o]], [[5xqk]] - EcTKT 1 + TPP + xylulose 5-P | + | **[[2r8o]], [[5xqk]] - EcTKT 1 + TPP + xylulose 5-P<br /> |
| - | + | **[[6tj9]] - EcTKT 1 + TPP analog + xylulose 5-P<br /> | |
**[[2r8p]] - EcTKT 1 + TPP + fructose 6-P<br /> | **[[2r8p]] - EcTKT 1 + TPP + fructose 6-P<br /> | ||
**[[5hje]] - PsTKT + TPP + seduheptulose 7-P<br /> | **[[5hje]] - PsTKT + TPP + seduheptulose 7-P<br /> | ||
| Line 77: | Line 83: | ||
**[[5xps]], [[5xu9]] - PsTKT + TPP + erythrose 4-P<br /> | **[[5xps]], [[5xu9]] - PsTKT + TPP + erythrose 4-P<br /> | ||
**[[5i5e]] - PsTKT (mutant) + TPP + xylulose 5-P<br /> | **[[5i5e]] - PsTKT (mutant) + TPP + xylulose 5-P<br /> | ||
| + | |||
| + | *Split-gene transketolase | ||
| + | |||
| + | **[[6yak]] - ChTKT N-terminal + C-terminal chain – ''Carboxydothermus hydrogenoformans''<br /> | ||
| + | **[[6yaj]] - ChTKT C-terminal chain<br /> | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 09:44, 14 December 2020
| |||||||||||
3D Structures of transketolase
Updated on 14-December-2020
References
- ↑ Verschueren KH, Kingma J, Rozeboom HJ, Kalk KH, Janssen DB, Dijkstra BW. Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide ions. Studies with halide compounds reveal a halide binding site in the active site. Biochemistry. 1993 Sep 7;32(35):9031-7. PMID:8369276
- ↑ James P, Isupov MN, De Rose SA, Sayer C, Cole IS, Littlechild JA. A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans: Structure and Biochemical Characterization. Front Microbiol. 2020 Oct 30;11:592353. doi: 10.3389/fmicb.2020.592353., eCollection 2020. PMID:33193259 doi:http://dx.doi.org/10.3389/fmicb.2020.592353
- ↑ Asztalos P, Parthier C, Golbik R, Kleinschmidt M, Hubner G, Weiss MS, Friedemann R, Wille G, Tittmann K. Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate. Biochemistry. 2007 Oct 30;46(43):12037-52. Epub 2007 Oct 3. PMID:17914867 doi:10.1021/bi700844m