This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1600
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
=bd oxidase; ''Geobacillus thermodenitrificans''= | =bd oxidase; ''Geobacillus thermodenitrificans''= | ||
| + | |||
==Structure== | ==Structure== | ||
<StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| + | |||
| + | == Structure == | ||
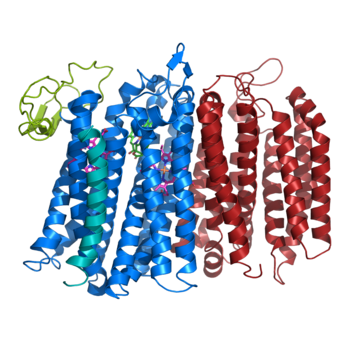
The overall structure contains 19 transmembrane helices that are arranged in a nearly oval shape. The protein contains two structurally similar subunits each containing nine helices (blue and red) and one smaller subunit, CydX, with one transmembrane helix. The subunits are interacting using hydrophobic residues and symmetry at the interfaces. The CydX subunit, whose function is not currently known, is positioned in the same way as CydS, which is found in E. coli bd oxidase. Due to its similar structure and position, it has been hypothesized to potentially stabilize heme b558 during potential structural rearrangements of the Q loop upon binding and oxidation of quinol. The Q loop is shown in lime green, and is a hydrophilic region above Cyd A. The lack of hydrogen bonding in this hydrophobic protein allows the protein to be flexible and go through a large conformational change for reduction of dioxygen. | The overall structure contains 19 transmembrane helices that are arranged in a nearly oval shape. The protein contains two structurally similar subunits each containing nine helices (blue and red) and one smaller subunit, CydX, with one transmembrane helix. The subunits are interacting using hydrophobic residues and symmetry at the interfaces. The CydX subunit, whose function is not currently known, is positioned in the same way as CydS, which is found in E. coli bd oxidase. Due to its similar structure and position, it has been hypothesized to potentially stabilize heme b558 during potential structural rearrangements of the Q loop upon binding and oxidation of quinol. The Q loop is shown in lime green, and is a hydrophilic region above Cyd A. The lack of hydrogen bonding in this hydrophobic protein allows the protein to be flexible and go through a large conformational change for reduction of dioxygen. | ||
[[Image:Bd oxidase structure (basic).png|350 px|right|thumb|Figure 1. bd oxidase; two structurally similar transmembrane helices in blue and red; CydX subunit in teal; Q loop in lime green. ]] | [[Image:Bd oxidase structure (basic).png|350 px|right|thumb|Figure 1. bd oxidase; two structurally similar transmembrane helices in blue and red; CydX subunit in teal; Q loop in lime green. ]] | ||
| - | |||
| - | == Function == | ||
| - | |||
== Disease == | == Disease == | ||
Revision as of 00:42, 23 March 2020
bd oxidase; Geobacillus thermodenitrificans
Structure
| |||||||||||