Coronavirus Disease 2019 (COVID-19)
From Proteopedia
(Difference between revisions)
| Line 64: | Line 64: | ||
* A team of scientists determined [http://www.rcsb.org/pdb/results/results.do?tabtoshow=Current&qrid=C212D599 '''25 crystal structure of SARS-CoV-2 main protease in complex with a series of different inhibitors''']. | * A team of scientists determined [http://www.rcsb.org/pdb/results/results.do?tabtoshow=Current&qrid=C212D599 '''25 crystal structure of SARS-CoV-2 main protease in complex with a series of different inhibitors''']. | ||
| - | * Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab<ref>PMID:32198291</ref>, Apo Struture: PDB-ID [http://www.rcsb.org/structure/6y2e 6Y2E], and complexes with inhibitors: PDB-ID [http://www.rcsb.org/structure/6y2f 6Y2F] and [http://www.rcsb.org/structure/6y2g 6Y2G]. | + | * Crystal structure of '''SARS-CoV-2 main protease''' provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab<ref>PMID:32198291</ref>, Apo Struture: PDB-ID [http://www.rcsb.org/structure/6y2e 6Y2E], and complexes with inhibitors: PDB-ID [http://www.rcsb.org/structure/6y2f 6Y2F] and [http://www.rcsb.org/structure/6y2g 6Y2G]. |
| - | * 3D Structure of RNA-dependent RNA polymerase from COVID-19, a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 [https://doi.org/10.1101/2020.03.16.993386 https://doi.org/10.1101/2020.03.16.993386]</ref>. | + | * 3D Structure of '''RNA-dependent RNA polymerase from COVID-19''', a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 [https://doi.org/10.1101/2020.03.16.993386 https://doi.org/10.1101/2020.03.16.993386]</ref>. |
| - | * Crystal structure of the Mpro from COVID-19 and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing <ref> Jin, et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.02.26.964882 http://doi.org/10.1101/2020.02.26.964882]</ref>, PDB-ID [[2h2z]]. | + | * Crystal structure of the '''Mpro from COVID-19''' and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing <ref> Jin, et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.02.26.964882 http://doi.org/10.1101/2020.02.26.964882]</ref>, PDB-ID [[2h2z]]. |
| - | * Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2 in a study by scientists from USA<ref> Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.02.968388 http://doi.org/10.1101/2020.03.02.968388]</ref>, PDB-ID [[6w01]]. | + | * Crystal structure of '''Nsp15 endoribonuclease NendoU from SARS-CoV-2''' in a study by scientists from USA<ref> Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.02.968388 http://doi.org/10.1101/2020.03.02.968388]</ref>, PDB-ID [[6w01]]. |
| - | * A study by Zhou & colleagues | + | * A study by Zhou & colleagues on the structural basis for the '''recognition of the SARS-CoV-2 (COVID-19) by full-length human ACE2''' gives insights to the molecular basis for coronavirus recognition and infection<ref>PMID:32132184</ref>. |
* '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]]. | * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]]. | ||
Revision as of 18:06, 25 March 2020
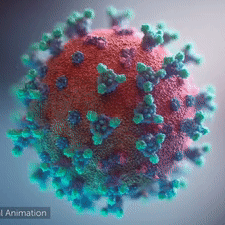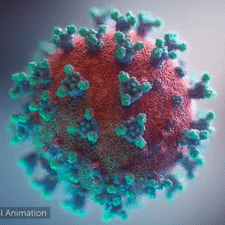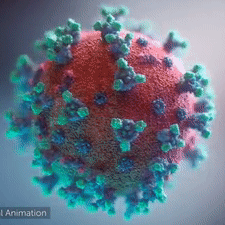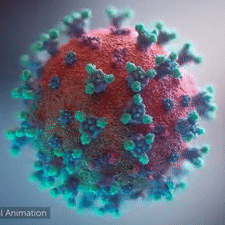
A novel coronavirus was found to be the cause of a respiratory illness first detected in Wuhan, China in 2019.
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Jaime Prilusky, Eric Martz, Jurgen Bosch, David Sehnal, Gianluca Santoni