This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1627
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ''Homo sapiens'' Insulin Receptor | |
<StructureSection load='6SOF' size='350' frame='true' side='right' caption='Insulin Receptor Ectodomain 6SOF' scene=''> | <StructureSection load='6SOF' size='350' frame='true' side='right' caption='Insulin Receptor Ectodomain 6SOF' scene=''> | ||
This is a default text for your page '''Harrison L. Smith/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Harrison L. Smith/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
| - | The insulin receptor is a transmembrane protein receptor that is a vital proponent of cellular function. It plays a key role in a variety of cellular pathways including glucose homeostasis, regulation of lipid, protein, and carbohydrate metabolism, gene expression, and even modulation of brain neurotransmitter levels. '''This page focuses specifically on the insulin receptor's role in glucose homeostasis'''. | + | The insulin receptor is a transmembrane protein receptor that is a vital proponent of cellular function. It plays a key role in a variety of cellular pathways including glucose homeostasis, regulation of lipid, protein, and carbohydrate metabolism, gene expression, and even modulation of brain neurotransmitter levels. '''This page focuses specifically on the insulin receptor's role in glucose homeostasis'''. |
==Structural Highlights== | ==Structural Highlights== | ||
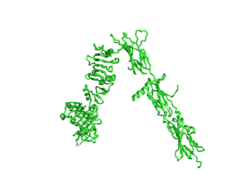
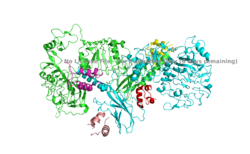
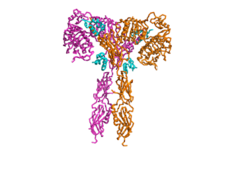
| - | The insulin receptor is a dimer of heterodimers made of two alpha subunits and two beta subunits <ref name=”Tatulian”>PMID:26322622</ref>. The <scene name='83/832953/Alpha_chain/1'>Alpha chain</scene> is on the extracellular side of the membrane and is critical for binding insulin. | + | The insulin receptor is a dimer of heterodimers made of two alpha subunits and two beta subunits <ref name=”Tatulian”>PMID:26322622</ref>. The insulin receptor in its most complete form can be viewed with <scene name='83/832953/Active_insulin_receptor/1'>four subunits and four binding sites</scene>. The <scene name='83/832953/Alpha_chain/1'>Alpha chain</scene> is on the extracellular side of the membrane and is critical for binding insulin. The <scene name='83/832953/Four_binding_site_locations/1'> binding sites locations</scene> that have the potential to interact with insulin on the extracellular side of the membrane, but it is generally more common for only one or two insulin molecules to bind to the receptor due to the occurrence of negative affinity at the binding site. The <scene name='83/832953/Beta_chains/1'>Beta chains</scene> are transmembrane subunits that contain a tyrosine kinase region. Beta chains experience a conformation change that brings them from a V shape to a T shape upon activation or binding of an insulin molecule. When the two subunits are brought near to each other in the activated T form, each Tyrosine Kinase region is able to autophosphorylate its counterparts at particular Tyrosine locations. This aspect of the molecule has not yet been imaged. |
== Function == | == Function == | ||
Revision as of 21:15, 29 March 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter