We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Harrison L. Smith/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ''Homo sapiens'' Insulin Receptor | |
| - | <StructureSection load='6SOF' size='350' frame='true' side='right' caption='Insulin Receptor Ectodomain 6SOF' scene=''> | + | <StructureSection load='6SOF' size='350' frame='true' side='right' caption='Insulin Receptor Ectodomain 6SOF' scene='83/832953/Active_insulin_receptor/1'> |
This is a default text for your page '''Harrison L. Smith/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Harrison L. Smith/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| Line 8: | Line 8: | ||
==Structural Highlights== | ==Structural Highlights== | ||
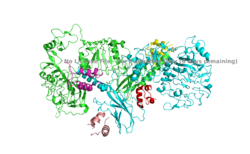
| - | The insulin receptor is a dimer of heterodimers made of two alpha subunits and two beta subunits <ref name=”Tatulian”>PMID:26322622</ref>. The <scene name='83/832953/ | + | The insulin receptor is a dimer of heterodimers made of two alpha subunits and two beta subunits <ref name=”Tatulian”>PMID:26322622</ref>.The <scene name='83/832953/Alpha_subunits/1'>Alpha chains</scene> are on the extracellular side of the membrane and is critical for binding insulin. The <scene name='83/832953/Binding_sites/1'>binding sites</scene> that have the potential to interact with insulin on the extracellular side of the membrane, but it is generally more common for only one or two insulin molecules to bind to the receptor due to the occurrence of negative affinity at the binding site. The <scene name='83/832953/Beta_subunits/1'>Beta chains</scene> are transmembrane subunits that contain a tyrosine kinase region. Beta chains experience a conformation change that brings them from a V shape to a T shape upon activation or binding of an insulin molecule. When the two subunits are brought near to each other in the activated T form, each Tyrosine Kinase region is able to autophosphorylate its counterparts at particular Tyrosine locations. This aspect of the molecule has not yet been imaged. |
| - | == Function | + | == Function == |
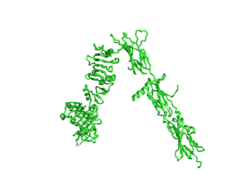
[[Image:Insulin inverted v.png|250px|right|thumb|Figure 1. An image of the inactive Insulin Receptor Alpha subunit in the inverted V conformation. This image shows only a protomer of the inactive alpha subunit because the entire inactive alpha subunit dimer has been unable to be photographed because the transition state has yet to be determined [https://www.rcsb.org/structure/6ce7/ PDB: 6CE7].]] | [[Image:Insulin inverted v.png|250px|right|thumb|Figure 1. An image of the inactive Insulin Receptor Alpha subunit in the inverted V conformation. This image shows only a protomer of the inactive alpha subunit because the entire inactive alpha subunit dimer has been unable to be photographed because the transition state has yet to be determined [https://www.rcsb.org/structure/6ce7/ PDB: 6CE7].]] | ||
| Line 21: | Line 21: | ||
The insulin receptor is a receptor tyrosine kinase that exists in two stable conformations, an inactive and active state. The entire insulin receptor is a dimer of heterodimers with two extracellular alpha subunits, and two transmembrane/intracellular beta subunits linked and stabilized by multiple disulfide bonds. | The insulin receptor is a receptor tyrosine kinase that exists in two stable conformations, an inactive and active state. The entire insulin receptor is a dimer of heterodimers with two extracellular alpha subunits, and two transmembrane/intracellular beta subunits linked and stabilized by multiple disulfide bonds. | ||
======Alpha Subunit====== | ======Alpha Subunit====== | ||
| - | The alpha subunit is the extracellular domain of the insulin receptor and is the site of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The actual site of insulin binding occurs at the | + | The alpha subunit is the extracellular domain of the insulin receptor and is the site of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The actual site of insulin binding occurs at the <scene name='83/832953/Alpha_c_helix/1'>α-CT chain</scene>. and is stabilized by the L1 and L2 domains. |
======Beta Subunit====== | ======Beta Subunit====== | ||
The beta subunit is the membrane spanning and intracellular portion of the insulin receptor. This domain is composed of three Fibronectin domains (FN III-1,-2,-3) and the tyrosine kinase domain. The tyrosine kinase domain is the site of autophosphorylation upon activation of the receptor. | The beta subunit is the membrane spanning and intracellular portion of the insulin receptor. This domain is composed of three Fibronectin domains (FN III-1,-2,-3) and the tyrosine kinase domain. The tyrosine kinase domain is the site of autophosphorylation upon activation of the receptor. | ||
| Line 28: | Line 28: | ||
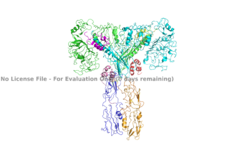
The insulin receptor can maximally bind four insulins to the active T-state at the four distinct insulin binding sites. Once insulin binds, the structural change from inactive to active state is stabilized by a tripartite interaction between insulin, L1, the C-terminal alpha helix, and the FNIII-1 domain. Studies have found that optimal insulin receptor activation requires the binding of multiple insulin ligands to two insulin binding sites. In (Figure 3) these two binding sites are colored in magenta and red. Binding of at least one insulin to the red binding site in (Figure 3) is required for the activation of the insulin receptor and the change in conformation to the active T state. <ref> DOI 10.7554/eLife.48630 </ref>. | The insulin receptor can maximally bind four insulins to the active T-state at the four distinct insulin binding sites. Once insulin binds, the structural change from inactive to active state is stabilized by a tripartite interaction between insulin, L1, the C-terminal alpha helix, and the FNIII-1 domain. Studies have found that optimal insulin receptor activation requires the binding of multiple insulin ligands to two insulin binding sites. In (Figure 3) these two binding sites are colored in magenta and red. Binding of at least one insulin to the red binding site in (Figure 3) is required for the activation of the insulin receptor and the change in conformation to the active T state. <ref> DOI 10.7554/eLife.48630 </ref>. | ||
| + | |||
| + | |||
| Line 34: | Line 36: | ||
=== Disease === | === Disease === | ||
One of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis is diabetes mellitus. There are two types of diabetes- which are referred to as type 1 and type 2 diabetes. Type 1 diabetes is classified as "insulin dependent" and is characterized by an inability for the body to produce insulin. This is most often the result of damage or insufficiency in the Islets of Langerhans in the pancreas. Type 2 diabetes is classified as "insulin independent" and is the result of the body producing insufficient amounts of insulin, or not responding to the insulin. This often occurs because of high blood-glucose levels. Both types of diabetes are often treated with insulin injections, and diet and lifestyle changes. <ref name="Wilcox"> PMID:16278749</ref> <ref name= "Riddle"> PMID: 6351440</ref>. | One of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis is diabetes mellitus. There are two types of diabetes- which are referred to as type 1 and type 2 diabetes. Type 1 diabetes is classified as "insulin dependent" and is characterized by an inability for the body to produce insulin. This is most often the result of damage or insufficiency in the Islets of Langerhans in the pancreas. Type 2 diabetes is classified as "insulin independent" and is the result of the body producing insufficient amounts of insulin, or not responding to the insulin. This often occurs because of high blood-glucose levels. Both types of diabetes are often treated with insulin injections, and diet and lifestyle changes. <ref name="Wilcox"> PMID:16278749</ref> <ref name= "Riddle"> PMID: 6351440</ref>. | ||
| + | ===At the Cellular Level=== | ||
| + | At the cellular level, the conformation change from the inactive to active state upon insulin binding has a time constant of six minutes. Once insulin binds and the beta subunits are brought within close proximity, autophosphorylation of the beta subunits begins. Phosphorylation at these sites reaches a maximal level in about one minute, and lasts for approximately six to ten minutes. One insulin receptor substrate has a half-life of 3.5 minutes where it is able to be phosphorylated by the tyrosine kinases of the beta subunit and then act as a central hub to activate further downstream signaling pathways that eventually bring glucose receptors to the surface of the cell to allow for diffusion of glucose into the cell. Once insulin binds to the alpha subunit, the receptor remains active for approximately ten minutes before the insulin is degraded and the receptor returns to its inactive conformation. | ||
[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1010832/ Treatment of Diabetes with Insulin] | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1010832/ Treatment of Diabetes with Insulin] | ||
Revision as of 22:39, 29 March 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter