Sandbox Reserved 1600
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
=Structure= | =Structure= | ||
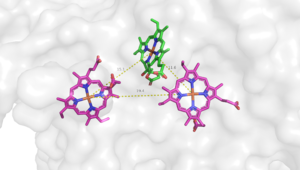
| - | The overall structure contains 19 transmembrane helices that are arranged in a nearly oval shape (Fig 1.) <ref name=”Safarian”>PMID: 27126043 </ref> The protein contains two structurally similar subunits each containing nine helices (blue and red) and one smaller subunit, CydX, with one transmembrane helix. The subunits are interacting using hydrophobic residues and symmetry at the interfaces. The CydX subunit, whose function is not currently known, is positioned in the same way as CydS, which is found in E. coli bd oxidase. Due to its similar structure and position, it has been hypothesized to potentially stabilize heme b558 during potential structural rearrangements of the Q loop upon binding and oxidation of quinol <ref name=”Safarian”>PMID: 27126043 </ref>. The Q loop is shown in lime green | + | The overall structure contains 19 transmembrane helices that are arranged in a nearly oval shape (Fig 1.) <ref name=”Safarian”>PMID: 27126043 </ref> The protein contains two structurally similar subunits each containing nine helices (blue and red) and one smaller subunit, CydX, with one transmembrane helix. The subunits are interacting using hydrophobic residues and symmetry at the interfaces. The CydX subunit, whose function is not currently known, is positioned in the same way as CydS, which is found in E. coli bd oxidase. Due to its similar structure and position, it has been hypothesized to potentially stabilize heme b558 during potential structural rearrangements of the Q loop upon binding and oxidation of quinol <ref name=”Safarian”>PMID: 27126043 </ref>. The Q loop is shown in lime green and is a hydrophilic region above Cyd A. The lack of hydrogen bonding in this hydrophobic protein allows the protein to be flexible and go through a large conformational change for reduction of dioxygen. |
==Active Site== | ==Active Site== | ||
| Line 19: | Line 19: | ||
==Potential Oxygen Entry Site== | ==Potential Oxygen Entry Site== | ||
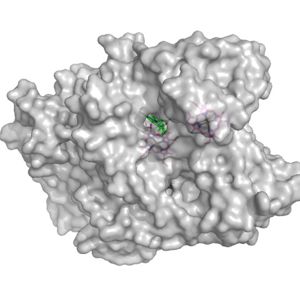
| - | [[Image:Potential_oxygen_entry_site.jpg|300 px|right|thumb|Figure 2. | + | [[Image:Potential_oxygen_entry_site.jpg|300 px|right|thumb|Figure 2. The surface of the potential oxygen entry site; Heme D showed in green]] |
Heme D is the hypothesized spot for the oxygen to enter the protein. Heme D is directly connected to the protein surface on CydA and contains a solvent accessible substrate channel (Fig 2.) | Heme D is the hypothesized spot for the oxygen to enter the protein. Heme D is directly connected to the protein surface on CydA and contains a solvent accessible substrate channel (Fig 2.) | ||
| Line 30: | Line 30: | ||
Because there is no proton pump present, the most likely proton transfer mechanism is facilitated by intracellular water molecules. | Because there is no proton pump present, the most likely proton transfer mechanism is facilitated by intracellular water molecules. | ||
| - | One potential proton pathway is formed from the four-helix bundle (a1-4) of CydA. It is called the CydA pathway. The location of Glu108 in our structure, together with previous mutagenesis experiments supports the proposal that this glutamate residue is a redox | + | One potential proton pathway is formed from the four-helix bundle (a1-4) of CydA. It is called the CydA pathway. The location of Glu108 in our structure, together with previous mutagenesis experiments supports the proposal that this glutamate residue is a redox state-dependent mediator of proton transfer to a charge compensation site. <ref name=”Safarian”>PMID: 27126043 </ref> With the CydA pathway leading to Glu101, this residue could be the protonatable group used for charge compensation upon heme b595 reduction. It is still unknown whether protons entering the CydA pathway can be transferred from Glu101 to Glu378, to allow the spread of the negative charge of a second electron used to reduce the two high-spin hemes. Proto- nation of Glu378 could alternatively also be accomplished by a proton accessing from the extracellular side. More research needs to be done to determine whether the CydA pathway is solely providing protons for charge compensation or whether Glu108 can be a branching point that is able to pass protons via the heme b595 propionates to the oxygen-binding site. |
Another potential entry site is related to the a1-4 four-helix bundle of CydB. Therefore, this is called the CydB pathway. In this pathway, Asp25 is thought to be the equivalent of the Glu108 in the CydA pathway <ref name=”Safarian”>PMID: 27126043 </ref>. The other residues help facilitate the movement of the proton very similarly to the CydA pathway. There is less known about the CydB pathway, and therefore, the CydA pathway is the most accepted source of protons. | Another potential entry site is related to the a1-4 four-helix bundle of CydB. Therefore, this is called the CydB pathway. In this pathway, Asp25 is thought to be the equivalent of the Glu108 in the CydA pathway <ref name=”Safarian”>PMID: 27126043 </ref>. The other residues help facilitate the movement of the proton very similarly to the CydA pathway. There is less known about the CydB pathway, and therefore, the CydA pathway is the most accepted source of protons. | ||
== Structure Similarity to Bd Oxidase found in ''Ecoli'' == | == Structure Similarity to Bd Oxidase found in ''Ecoli'' == | ||
| - | [[Image:Aligmentbdoidase.jpg| | + | The structure of Bd Oxidase for ''Geobacillus thermodenitrificans'' is highly similar to the structure of Bd Oxidase for ''Ecoli'' with the only noticeable difference being the length of the Q-loop. The similarity and differences between the two proteins can be seen in the alignment of their main structures (Fig.4). [[Image:Aligmentbdoidase.jpg|200 px|right|thumb|Figure 4. Alignment of bd oxidase for the organisms ''Geobacillus thermodenitrificans'' (blue) and ''Ecoli'' (purple).]] Although only having one noticeable difference in structure, this difference causes the two proteins to have different active sites. In particular, the hemes of bd oxidase in ''Ecoli'' are arranged differently than the hemes in ''Geobacillus thermodenitrificans''. The main reason for this change in heme arrangement is because of the oxygen-binding site being blocked by the Q-loop in ''Ecoli''. |
Revision as of 00:44, 31 March 2020
bd oxidase; Geobacillus thermodenitrificans
| |||||||||||
References
- ↑ Giuffre A, Borisov VB, Arese M, Sarti P, Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014 Jul;1837(7):1178-87. doi:, 10.1016/j.bbabio.2014.01.016. Epub 2014 Jan 31. PMID:24486503 doi:http://dx.doi.org/10.1016/j.bbabio.2014.01.016
- ↑ Safarian S, Hahn A, Mills DJ, Radloff M, Eisinger ML, Nikolaev A, Meier-Credo J, Melin F, Miyoshi H, Gennis RB, Sakamoto J, Langer JD, Hellwig P, Kuhlbrandt W, Michel H. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science. 2019 Oct 4;366(6461):100-104. doi: 10.1126/science.aay0967. PMID:31604309 doi:http://dx.doi.org/10.1126/science.aay0967
- ↑ Junemann S. Cytochrome bd terminal oxidase. Biochim Biophys Acta. 1997 Aug 22;1321(2):107-27. doi:, 10.1016/s0005-2728(97)00046-7. PMID:9332500 doi:http://dx.doi.org/10.1016/s0005-2728(97)00046-7
- ↑ Das A, Silaghi-Dumitrescu R, Ljungdahl LG, Kurtz DM Jr. Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J Bacteriol. 2005 Mar;187(6):2020-9. doi: 10.1128/JB.187.6.2020-2029.2005. PMID:15743950 doi:http://dx.doi.org/10.1128/JB.187.6.2020-2029.2005
- ↑ Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477