This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1627
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
The beta subunit is the membrane spanning and intracellular portion of the insulin receptor. This domain is composed of three Fibronectin domains (FN III-1,-2,-3) and the tyrosine kinase domain. The tyrosine kinase domain is the site of autophosphorylation upon activation of the receptor. | The beta subunit is the membrane spanning and intracellular portion of the insulin receptor. This domain is composed of three Fibronectin domains (FN III-1,-2,-3) and the tyrosine kinase domain. The tyrosine kinase domain is the site of autophosphorylation upon activation of the receptor. | ||
===Conformation Change=== | ===Conformation Change=== | ||
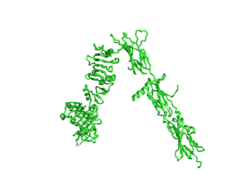
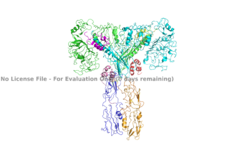
| - | The inactive form of the insulin receptor predominates in low-levels of circulating insulin, whereas the active conformation is seen when insulin binds to any of the 4 receptor sites. The inactive conformation resembles an inverted V (Figure 1), and the active conformation resembles a T (Figure | + | The inactive form of the insulin receptor predominates in low-levels of circulating insulin, whereas the active conformation is seen when insulin binds to any of the 4 receptor sites. The inactive conformation resembles an inverted V (Figure 1), and the active conformation resembles a T (Figure 3). Upon the binding of insulin to any of the four binding sites, the conformation change will begin, causing the Beta subunit's tyrosine kinase domains to move close together, allowing them to autophosphorylate. This autophosphorylation is what activates the insulin receptor and allows it to participate in further downstream signaling pathways. <ref> DOI 10.1038/s41467-018-06826-6</ref>. |
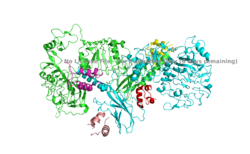
The insulin receptor can maximally bind four insulins to the active T-state at the four distinct insulin binding sites. Once insulin binds, the structural change from inactive to active state is stabilized by a tripartite interaction between insulin, L1, the C-terminal alpha helix, and the FNIII-1 domain. Studies have found that optimal insulin receptor activation requires the binding of multiple insulin ligands to two insulin binding sites. In (Figure 3) these two binding sites are colored in magenta and red. Binding of at least one insulin to the red binding site in (Figure 3) is required for the activation of the insulin receptor and the change in conformation to the active T state. <ref> DOI 10.7554/eLife.48630 </ref>. | The insulin receptor can maximally bind four insulins to the active T-state at the four distinct insulin binding sites. Once insulin binds, the structural change from inactive to active state is stabilized by a tripartite interaction between insulin, L1, the C-terminal alpha helix, and the FNIII-1 domain. Studies have found that optimal insulin receptor activation requires the binding of multiple insulin ligands to two insulin binding sites. In (Figure 3) these two binding sites are colored in magenta and red. Binding of at least one insulin to the red binding site in (Figure 3) is required for the activation of the insulin receptor and the change in conformation to the active T state. <ref> DOI 10.7554/eLife.48630 </ref>. | ||
Revision as of 01:01, 30 March 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter