We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1616
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
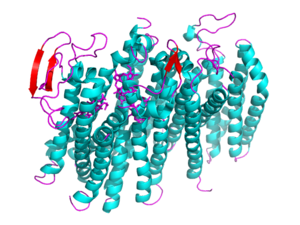
[[Image: 5doq_WHOLE_IMAGE.png|300 px|left|thumb|Figure 2: 6RX4 monomer subunit; alpha helices in teal, beta sheets in purple.]] | [[Image: 5doq_WHOLE_IMAGE.png|300 px|left|thumb|Figure 2: 6RX4 monomer subunit; alpha helices in teal, beta sheets in purple.]] | ||
{{Clear}} | {{Clear}} | ||
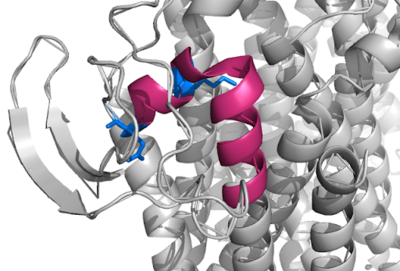
| - | This page will be specifically focusing on the structure and overall function of the 6RX4 bd oxidase. 6RX4 is a part of the long(L) quinol-binding domain subfamily that terminal oxidases are classified into. The L-subfamily of bd oxidases are responsible for the survival of acute infectious diseases such as E.Coli and salmonella. The 6RX4's three <scene name='83/832931/Heme/4'>heme</scene> groups, its periplasmically exposed Q loop, and four protein subunits will be of primary focus when identifying the relationship between structure and function. | + | This page will be specifically focusing on the structure and overall function of the 6RX4 bd oxidase. 6RX4 is a part of the long(L) quinol-binding domain subfamily that terminal oxidases are classified into. The L-subfamily of bd oxidases are responsible for the survival of acute infectious diseases such as E.Coli and salmonella. The 6RX4's three <scene name='83/832931/Heme/4'>heme</scene> groups, its periplasmically exposed <scene name='83/832924/Q_loop/3'>Q-loop</scene>, and four protein subunits will be of primary focus when identifying the relationship between structure and function. |
== Function == | == Function == | ||
| Line 23: | Line 23: | ||
== Structural highlights == | == Structural highlights == | ||
[[Image:Q Loop.png|400 px|right|thumb|Figure 1]] | [[Image:Q Loop.png|400 px|right|thumb|Figure 1]] | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | <scene name='83/837229/Residues_22-44_of_bd_oxidase/2'>bd Oxidase Residues 22-44</scene> | ||
Revision as of 04:18, 31 March 2020
bd Oxidase, 5DOQ
| |||||||||||
References
- ↑ 1.0 1.1 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ 2.0 2.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
![Figure 1: Overall schematic representation of cytochrome bd; General display of the reduction of molecular oxygen into water [1]](/wiki/images/thumb/e/e6/Proton_graadient.jpg/300px-Proton_graadient.jpg)