We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1616
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
bd Oxidase is a type of quinol-dependent terminal oxidase found exclusively in prokaryotes. With a very high oxygen affinity, bd oxidases play a vital role in the oxidative phosphorylation pathway in both gram-positive and gram-negative bacteria. bd oxidases responsibility in the oxidative phosphorylation pathway allows the protein to also assist as a key survival factor in the bacterial stress response against antibacterial drugs. Given this knowledge, bd oxidases have become an area of scientific research worth pursuing as they could serve as an ideal target for antimicrobial drug development. | bd Oxidase is a type of quinol-dependent terminal oxidase found exclusively in prokaryotes. With a very high oxygen affinity, bd oxidases play a vital role in the oxidative phosphorylation pathway in both gram-positive and gram-negative bacteria. bd oxidases responsibility in the oxidative phosphorylation pathway allows the protein to also assist as a key survival factor in the bacterial stress response against antibacterial drugs. Given this knowledge, bd oxidases have become an area of scientific research worth pursuing as they could serve as an ideal target for antimicrobial drug development. | ||
| - | [[Image:proton graadient.jpg|300 px|left|thumb|Figure 1: Overall schematic representation of cytochrome bd; General display of the reduction of molecular oxygen into water [https://doi.org/10.1016/j.bbabio.2014.01.016.]]] | + | [[Image:proton graadient.jpg|300 px|left|thumb|Figure 1: Overall schematic representation of cytochrome bd; General display of the reduction of molecular oxygen into water using the quinol as a reducing substrate. [https://doi.org/10.1016/j.bbabio.2014.01.016.]]] |
{{Clear}} | {{Clear}} | ||
| - | The overall mechanism of bd oxidases involves an exergonic reduction reaction of molecular oxygen into water. During this reaction, a proton gradient is generated in order to assist in the conservation of energy. Unlike other terminal oxidases, bd oxidases do not use a proton pump. Instead, bd oxidases use a form of vectorial chemistry that releases protons from the quinol oxidation into the positive, periplasmic side of the membrane. Protons that are required for the water formation are then consumed from the negative, cytoplasmic side of the membrane, thus creating the previously mentioned proton gradient. | + | The overall mechanism of bd oxidases involves an exergonic reduction reaction of molecular oxygen into water (Figure 1). During this reaction, a proton gradient is generated in order to assist in the conservation of energy. Unlike other terminal oxidases, bd oxidases do not use a proton pump. Instead, bd oxidases use a form of vectorial chemistry that releases protons from the quinol oxidation into the positive, periplasmic side of the membrane. Protons that are required for the water formation are then consumed from the negative, cytoplasmic side of the membrane, thus creating the previously mentioned proton gradient. |
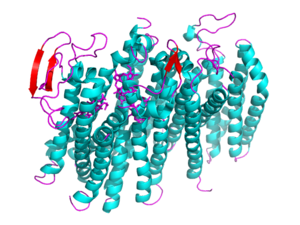
[[Image: 5doq_WHOLE_IMAGE.png|300 px|left|thumb|Figure 2: 6RX4 monomer subunit; alpha helices in teal, beta sheets in purple.]] | [[Image: 5doq_WHOLE_IMAGE.png|300 px|left|thumb|Figure 2: 6RX4 monomer subunit; alpha helices in teal, beta sheets in purple.]] | ||
| Line 22: | Line 22: | ||
== Structural highlights == | == Structural highlights == | ||
| - | [[Image:Q Loop.png|400 px|right|thumb|Figure 1]] | ||
Revision as of 05:03, 31 March 2020
bd Oxidase, 5DOQ
| |||||||||||
References
- ↑ 1.0 1.1 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ 2.0 2.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
![Figure 1: Overall schematic representation of cytochrome bd; General display of the reduction of molecular oxygen into water using the quinol as a reducing substrate. [1]](/wiki/images/thumb/e/e6/Proton_graadient.jpg/300px-Proton_graadient.jpg)