We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1626
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
=== Selectivity Filter === | === Selectivity Filter === | ||
| - | The pore-forming subunit of the MCU contains 351 amino acid residues with both the N- and C-terminal domains located in the matrix of the mitochondria. The two transmembrane domains, TM1 and TM2, are connected by a solvent- exposed loop with a highly conserved <scene name='83/832952/Starting_scene/ | + | The pore-forming subunit of the MCU contains 351 amino acid residues with both the N- and C-terminal domains located in the matrix of the mitochondria. The two transmembrane domains, TM1 and TM2, are connected by a solvent- exposed loop with a highly conserved <scene name='83/832952/Starting_scene/3'>DXXE motif</scene> which is essential for the calcium transport, located in the upper helix of TM2. The first pore-lining residues are <scene name='83/832952/Selectivity_filter_asp/1'>Asp333</scene> and <scene name='83/832952/Selectivity_filter_glu/1'>Glu336</scene>, that are both part of the highly conserved DXXE motif connecting TM1 and TM2. Each monomer has a Glu336 whose carboxylate group points toward the pore center. The diameter of the ring is 5Å meaning that the calcium is dehydrated. Trp332 stabilizes the carboxyl groups of two neighboring Glu336 residues through hydrogen bonding. The additional interaction of Trp332 with Pro337 serves to orient Glu336 for calcium coordination. Therefore, Glu336, Trp332, and Pro337 make up the highly conserved selectivity filter. <ref name="Fan"/> |
=== Common Mutations === | === Common Mutations === | ||
| Line 40: | Line 40: | ||
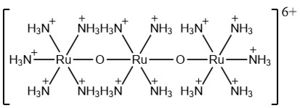
Inorganic salts and coordination complexes have also been shown to inhibit calcium uptake. Specifically, the trivalent lanthanide ions can competitively inhibit the uniporter because of their similar ionic radii and coordination preferences to calcium. In addition, several transition metal coordination complexes (most notably Co, Cr, and Rh) with amine ligands have been shown to inhibit calcium uptake. Again, there is no apparent structure-activity relationship that predicts this behavior.<ref name="Woods" /> | Inorganic salts and coordination complexes have also been shown to inhibit calcium uptake. Specifically, the trivalent lanthanide ions can competitively inhibit the uniporter because of their similar ionic radii and coordination preferences to calcium. In addition, several transition metal coordination complexes (most notably Co, Cr, and Rh) with amine ligands have been shown to inhibit calcium uptake. Again, there is no apparent structure-activity relationship that predicts this behavior.<ref name="Woods" /> | ||
| - | The most well-known and commonly used inhibitor of calcium uptake into the mitochondria is ruthenium red ([https://en.wikipedia.org/wiki/Ruthenium_red RuRed]). [[Image:Ruthenium_Red.jpg|300 px|right|thumb|'''Fig. 2''' Structure of Ruthenium Red]] RuRed effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. Additionally, it has been shown to mitigate tissue damage due to IRI and slow cancer cell migration. The issue with RuRed is that its purification has always been a challenging matter.<ref name="Woods" /> Interestingly enough, this led to even more developments in the search for an inhibitor. Many scientists had observed that impure RuRed actually had greater inhibition than pure RuRed. One of the common minor impurities of RuRed, [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the active component of the RuRed mixtures, meaning it responsible for calcium inhibition. Ru360 is now commercially available and has been widely used for the study of calcium-dependent cellular processes and as a therapeutic agent. Very little is known about its mechanism of inhibtion, but studies show that it interacts with the <scene name='83/832952/Starting_scene/ | + | The most well-known and commonly used inhibitor of calcium uptake into the mitochondria is ruthenium red ([https://en.wikipedia.org/wiki/Ruthenium_red RuRed]). [[Image:Ruthenium_Red.jpg|300 px|right|thumb|'''Fig. 2''' Structure of Ruthenium Red]] RuRed effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. Additionally, it has been shown to mitigate tissue damage due to IRI and slow cancer cell migration. The issue with RuRed is that its purification has always been a challenging matter.<ref name="Woods" /> Interestingly enough, this led to even more developments in the search for an inhibitor. Many scientists had observed that impure RuRed actually had greater inhibition than pure RuRed. One of the common minor impurities of RuRed, [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the active component of the RuRed mixtures, meaning it responsible for calcium inhibition. Ru360 is now commercially available and has been widely used for the study of calcium-dependent cellular processes and as a therapeutic agent. Very little is known about its mechanism of inhibtion, but studies show that it interacts with the <scene name='83/832952/Starting_scene/3'>DXXE motif</scene> of the loop connecting the TM1 and TM2 helices.<ref name="Woods" /> |
Ru360 was a very successful inhibitor, but it showed low cell permeability. So, a new inhibitor called Ru265 was developed which could be easily synthesized and didn't need chromatographic purification. Ru265 had all of the benefits of Ru360, with with twice the cell permeability. Additionally, the same mutations didn't seem to affect it. Mutations of D261 and S259 in human MCU reduced inhibitory effect of Ru360, but not Ru265. Additionally, there were other mutations that affected Ru265, but not Ru360.<ref name="Woods" /> This shows how much more research is needed before a mechanism is understood for any inhibitor of the MCU. | Ru360 was a very successful inhibitor, but it showed low cell permeability. So, a new inhibitor called Ru265 was developed which could be easily synthesized and didn't need chromatographic purification. Ru265 had all of the benefits of Ru360, with with twice the cell permeability. Additionally, the same mutations didn't seem to affect it. Mutations of D261 and S259 in human MCU reduced inhibitory effect of Ru360, but not Ru265. Additionally, there were other mutations that affected Ru265, but not Ru360.<ref name="Woods" /> This shows how much more research is needed before a mechanism is understood for any inhibitor of the MCU. | ||
Revision as of 19:27, 5 April 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
Mitochondrial Calcium Uniporter (MCU) Complex
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 2.0 2.1 2.2 2.3 2.4 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 3.0 3.1 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
- ↑ Kamer KJ, Jiang W, Kaushik VK, Mootha VK, Grabarek Z. Crystal structure of MICU2 and comparison with MICU1 reveal insights into the uniporter gating mechanism. Proc Natl Acad Sci U S A. 2019 Feb 12. pii: 1817759116. doi:, 10.1073/pnas.1817759116. PMID:30755530 doi:http://dx.doi.org/10.1073/pnas.1817759116
- ↑ Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
Student Contributors
Ryan Heumann
Rieser Wells