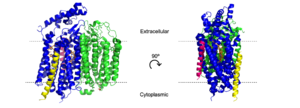We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1625
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
=== Subunits === | === Subunits === | ||
| - | [https://en.wikipedia.org/wiki/Escherichia_coli ''E. coli''] bd oxidase is made up of four individual subunits.<ref name="Alexander">PMID:31723136</ref> The two major subunits, CydA and CydB, are each composed of one peripheral helix and two bundles of four transmembrane helices. The <scene name='83/832924/Cyda_subunit/6'>CydA subunit</scene> plays the most important role in the oxygen reduction reaction as it contains the Q-loop as well as all three heme groups. The <scene name='83/832924/Cydb_subunit/2'>CydB subunit</scene> harbors the <scene name='83/832924/Ubiquinone/3'>ubiquinone</scene> molecule which provides structural support to the subunit that mimics the three hemes found in CydA.<ref name="Safarian">PMID: 31604309</ref><ref name="Safarian2">PMID: 27126043</ref> The remaining two subunits, CydS and CydX, are both single helix structures that assist in the oxygen reduction reaction. Unique to ''E. coli'', the <scene name='83/832924/Cyds_subunit/ | + | [https://en.wikipedia.org/wiki/Escherichia_coli ''E. coli''] bd oxidase is made up of four individual subunits.<ref name="Alexander">PMID:31723136</ref> The two major subunits, CydA and CydB, are each composed of one peripheral helix and two bundles of four transmembrane helices. The <scene name='83/832924/Cyda_subunit/6'>CydA subunit</scene> plays the most important role in the oxygen reduction reaction as it contains the Q-loop as well as all three heme groups. The <scene name='83/832924/Cydb_subunit/2'>CydB subunit</scene> harbors the <scene name='83/832924/Ubiquinone/3'>ubiquinone</scene> molecule which provides structural support to the subunit that mimics the three hemes found in CydA.<ref name="Safarian">PMID: 31604309</ref><ref name="Safarian2">PMID: 27126043</ref> The remaining two subunits, CydS and CydX, are both single helix structures that assist in the oxygen reduction reaction. Unique to ''E. coli'', the <scene name='83/832924/Cyds_subunit/5'>CydS subunit</scene> binds to CydA to block oxygen from directly binding to heme b595. The <scene name='83/832924/Cydx_subunit/5'>CydX subunit</scene> promotes the assembly and stability of the oxidase complex. CydX is composed of 37 mostly hydrophilic amino acid residues, including <scene name='83/832924/Glu25/2'>Glu25</scene> that is exposed to the cytoplasm and prevents the helix from fully entering the membrane. <ref name="Alexander">PMID:31723136</ref> |
===Q-Loop=== | ===Q-Loop=== | ||
Revision as of 02:46, 7 April 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
Cytochrome bd-1 oxidase in Escherichia coli
| |||||||||||
References
- ↑ 1.0 1.1 Harikishore A, Chong SSM, Ragunathan P, Bates RW, Gruber G. Targeting the menaquinol binding loop of mycobacterial cytochrome bd oxidase. Mol Divers. 2020 Jan 14. pii: 10.1007/s11030-020-10034-0. doi:, 10.1007/s11030-020-10034-0. PMID:31939065 doi:http://dx.doi.org/10.1007/s11030-020-10034-0
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Thesseling A, Rasmussen T, Burschel S, Wohlwend D, Kagi J, Muller R, Bottcher B, Friedrich T. Homologous bd oxidases share the same architecture but differ in mechanism. Nat Commun. 2019 Nov 13;10(1):5138. doi: 10.1038/s41467-019-13122-4. PMID:31723136 doi:http://dx.doi.org/10.1038/s41467-019-13122-4
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 Safarian S, Hahn A, Mills DJ, Radloff M, Eisinger ML, Nikolaev A, Meier-Credo J, Melin F, Miyoshi H, Gennis RB, Sakamoto J, Langer JD, Hellwig P, Kuhlbrandt W, Michel H. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science. 2019 Oct 4;366(6461):100-104. doi: 10.1126/science.aay0967. PMID:31604309 doi:http://dx.doi.org/10.1126/science.aay0967
- ↑ 4.0 4.1 4.2 4.3 4.4 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ 5.0 5.1 Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Buttner L, Smoot MP, Behrendt CL, Cherry S, Santos RL, Hooper LV, Winter SE. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017 Feb 8;21(2):208-219. doi: 10.1016/j.chom.2017.01.005. PMID:28182951 doi:http://dx.doi.org/10.1016/j.chom.2017.01.005
- ↑ 6.0 6.1 Shepherd M, Achard ME, Idris A, Totsika M, Phan MD, Peters KM, Sarkar S, Ribeiro CA, Holyoake LV, Ladakis D, Ulett GC, Sweet MJ, Poole RK, McEwan AG, Schembri MA. The cytochrome bd-I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci Rep. 2016 Oct 21;6:35285. doi: 10.1038/srep35285. PMID:27767067 doi:http://dx.doi.org/10.1038/srep35285
- ↑ Arora K, Ochoa-Montano B, Tsang PS, Blundell TL, Dawes SS, Mizrahi V, Bayliss T, Mackenzie CJ, Cleghorn LA, Ray PC, Wyatt PG, Uh E, Lee J, Barry CE 3rd, Boshoff HI. Respiratory flexibility in response to inhibition of cytochrome C oxidase in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014 Nov;58(11):6962-5. doi: 10.1128/AAC.03486-14., Epub 2014 Aug 25. PMID:25155596 doi:http://dx.doi.org/10.1128/AAC.03486-14
- ↑ Galvan AE, Chalon MC, Rios Colombo NS, Schurig-Briccio LA, Sosa-Padilla B, Gennis RB, Bellomio A. Microcin J25 inhibits ubiquinol oxidase activity of purified cytochrome bd-I from Escherichia coli. Biochimie. 2019 May;160:141-147. doi: 10.1016/j.biochi.2019.02.007. Epub 2019 Feb, 19. PMID:30790617 doi:http://dx.doi.org/10.1016/j.biochi.2019.02.007
- ↑ Lu P, Heineke MH, Koul A, Andries K, Cook GM, Lill H, van Spanning R, Bald D. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep. 2015 May 27;5:10333. doi: 10.1038/srep10333. PMID:26015371 doi:http://dx.doi.org/10.1038/srep10333
Student Contributors
- Grace Bassler
- Emily Neal
- Marisa Villarreal