We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1607
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Structural highlights and mechanism == | == Structural highlights and mechanism == | ||
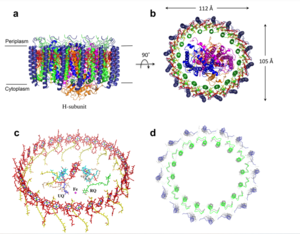
| - | The MCU is a dimer of dimers, described as tetrameric truncated pyramid. The uniporter has only a single strong binding site located in the selectivity pore with specificity for [https://en.wikipedia.org/wiki/Calcium_signaling Calcium], near the surface of the inner mitochondrial membrane. <ref> DOI: 10.1038/s41586-018-0330-9</ref> Activity of the uniporter is dependent on membrane potential and calcium concentration. Calcium from the cytoplasm enters the mitochondrial inner membrane space through the mitochondrial membrane and is passed to the mitochondrial matrix via the MCU. [[Image:structure.png|300 px|right|thumb|Figure 2: structure of mitochondrial calcium uniporter]] | + | The MCU is a dimer of dimers, described as tetrameric truncated pyramid. The uniporter has only a single strong binding site located in the selectivity pore with specificity for [https://en.wikipedia.org/wiki/Calcium_signaling Calcium], near the surface of the inner mitochondrial membrane. <ref name="Fan C"> DOI: 10.1038/s41586-018-0330-9</ref> Activity of the uniporter is dependent on membrane potential and calcium concentration. Calcium from the cytoplasm enters the mitochondrial inner membrane space through the mitochondrial membrane and is passed to the mitochondrial matrix via the MCU. [[Image:structure.png|300 px|right|thumb|Figure 2: structure of mitochondrial calcium uniporter]] |
===Transmembrane Domain=== | ===Transmembrane Domain=== | ||
| - | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space. The small pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) while <scene name='83/837230/Transmembrane_1/2'>transmembrane 1</scene> (TM1) surrounds the pore. The transmembrane domain exhibits four fold rotational symmetry. It is important that the selectivity pore is small, allowing only a dehydrated calcium molecule to interact with the 5 ampier wide glutamate ring. The negative charge of the glutamates carboxyl group attracts the positively charged Calcium ion. Approximately one helical turn below the glutamate ring of the selectivity filter, there is a tyrosine ring coming a 12 ampier wide pore allowing high conductivity. <ref | + | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space. The small pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) while <scene name='83/837230/Transmembrane_1/2'>transmembrane 1</scene> (TM1) surrounds the pore. The transmembrane domain exhibits four fold rotational symmetry. It is important that the selectivity pore is small, allowing only a dehydrated calcium molecule to interact with the 5 ampier wide glutamate ring. The negative charge of the glutamates carboxyl group attracts the positively charged Calcium ion. Approximately one helical turn below the glutamate ring of the selectivity filter, there is a tyrosine ring coming a 12 ampier wide pore allowing high conductivity. <ref name="Fan C" /> The wider opening allows calcium to rehydrate once they pass the selectivity pore. The domain swapping of TM1 of one subunit with the TM2 of the neighboring subunits allows for a tight packing in the transmembrane connectivity providing flexibility to the uniporter. |
===Soluble Domain=== | ===Soluble Domain=== | ||
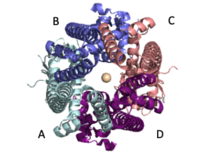
| - | The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Calcium ions down their concentration gradient.<ref | + | The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Calcium ions down their concentration gradient.<ref name="Fan C" /> The junction between the transmembrane domain and the coiled coil's flexibility can be attributed to the disordered packing between subunits; subunits A and C adopt different conformations than the B and D subunits, although they superimpose well.<ref name="Fan C" /> [[Image:Symmetry.png|200 px|left|thumb|Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane]]When calcium binds to the selectivity pore, the coiled coil swings approximately 8 degrees around its end near the <scene name='83/837230/Coiled_coil/3'>N-terminal domain</scene>. This movement propagates to the top of the transmembrane domain, where the pore is located, about 85 amperes away. The largest displacement triggered by the movement of the coiled coil is in the transmembrane domain, where the coil bends 20 degrees, moving the transmembrane domain further apart. The N-Terminal domain (NTD) is involved in calcium condition. Reorganization in the NTD due to shifts in the coiled coil switch subunits to alter membrane packing causing the interactions between the tyrosines and transmembrane helices. This propagation facilitates a rotamer switch between one pair of tyrosine controlling calcium flow through the pore. The soluble domain is wider than the transmembrane domain, allowing calcium ions to rehydrate and increasing the conductivity of ions through the uniporter into the mitochondrial matrix.<ref name="Fan C" /> |
| - | + | ||
===Selectivity Filter=== | ===Selectivity Filter=== | ||
Revision as of 04:39, 7 April 2020
Mitochondrial Calcium Uniporter
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.0 4.1 4.2 4.3 4.4 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
- ↑ Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018 Jun 28. pii: science.aar4056. doi: 10.1126/science.aar4056. PMID:29954988 doi:http://dx.doi.org/10.1126/science.aar4056
- ↑ Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8