We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1606
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
===ATP Bound and Unbound Conformations=== | ===ATP Bound and Unbound Conformations=== | ||
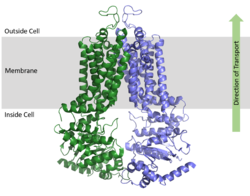
| - | As an [https://en.wikipedia.org/wiki/ATP-binding_cassette_transporter ABC Transporter], ABCG2 exhibits ATPase activity, using the energy of ATP hydrolysis to facilitate transport. After substrate bind in the TMD, one molecule of <scene name='83/832932/Atp_bound_use2/3'>ATP binds each NBD</scene> (2 molecules of ATP total) causing a conformational change of the overall structure from an <scene name='83/832932/Overall_use_2/3'>inward-facing conformation</scene> to an <scene name='83/832932/Outward_facing_conformation/4'>outward-facing conformation</scene>. One molecule of ATP is hydrolyzed to transport substrates across the cell membrane while the second molecule of ATP is hydrolyzed to reset the transporter to its inward-facing conformation.<ref name="Robey"/> | + | As an [https://en.wikipedia.org/wiki/ATP-binding_cassette_transporter ABC Transporter], ABCG2 exhibits ATPase activity, using the energy of ATP hydrolysis to facilitate transport. After substrate bind in the TMD, one molecule of <scene name='83/832932/Atp_bound_use2/3'>ATP binds each NBD</scene> (2 molecules of ATP total) causing a conformational change of the overall structure from an <scene name='83/832932/Overall_use_2/3'>inward-facing conformation</scene> to an <scene name='83/832932/Outward_facing_conformation/4'>outward-facing conformation</scene>. ATP coordinates with various residues and a magnesium ion in the <scene name='83/832932/Atp_bound_in_nbd/2'>binding site of each NBD</scene>. One molecule of ATP is hydrolyzed to transport substrates across the cell membrane while the second molecule of ATP is hydrolyzed to reset the transporter to its inward-facing conformation.<ref name="Robey"/> |
When ATP binds, α-helices in the NBD <scene name='83/832932/Atp_bound_nbd/3'>rotate</scene> approximately 35° relative to the <scene name='83/832932/Overall_structure_nbd_unbound/5'>inward-facing conformation of NBD</scene>. This shift in the NBD causes slight shifts of α-helices in the TMD; these helices are <scene name='83/832932/Atp_bound_use_tmd/3'>pushed toward each other</scene> relative to the <scene name='83/832932/Overall_structure_tmd_unbound/4'>inward-facing conformation of TMD</scene>. The overall shift from inward-facing to outward-facing promotes the transport of substrates through the transporter.<ref name="Manolaridis"/> | When ATP binds, α-helices in the NBD <scene name='83/832932/Atp_bound_nbd/3'>rotate</scene> approximately 35° relative to the <scene name='83/832932/Overall_structure_nbd_unbound/5'>inward-facing conformation of NBD</scene>. This shift in the NBD causes slight shifts of α-helices in the TMD; these helices are <scene name='83/832932/Atp_bound_use_tmd/3'>pushed toward each other</scene> relative to the <scene name='83/832932/Overall_structure_tmd_unbound/4'>inward-facing conformation of TMD</scene>. The overall shift from inward-facing to outward-facing promotes the transport of substrates through the transporter.<ref name="Manolaridis"/> | ||
| - | <scene name='83/832932/Atp_bound_in_nbd/2'>Text To Be Displayed</scene> | ||
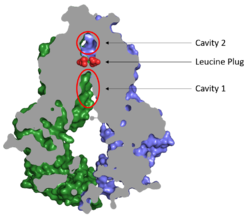
===Cavities and Leucine Plug=== | ===Cavities and Leucine Plug=== | ||
[[Image:Map_leucine_plug.png|250 px|right|thumb|Figure 2. Locations of Cavities 1 and 2 and the Leucine Plug in ABCG2. The protein is in the inward-facing conformation with Cavity 1 open to the cytosol for substrate recruitment, the Leucine Plug is intact, and Cavity 2 is completely occluded. [https://www.rcsb.org/structure/5NJ3 (5NJ3)]]] | [[Image:Map_leucine_plug.png|250 px|right|thumb|Figure 2. Locations of Cavities 1 and 2 and the Leucine Plug in ABCG2. The protein is in the inward-facing conformation with Cavity 1 open to the cytosol for substrate recruitment, the Leucine Plug is intact, and Cavity 2 is completely occluded. [https://www.rcsb.org/structure/5NJ3 (5NJ3)]]] | ||
Revision as of 01:39, 17 April 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
ABCG2 Multidrug Transporter
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017 Jun 22;546(7659):504-509. doi: 10.1038/nature22345. Epub 2017 May, 29. PMID:28554189 doi:http://dx.doi.org/10.1038/nature22345
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018 Nov;563(7731):426-430. doi: 10.1038/s41586-018-0680-3. Epub 2018 Nov, 7. PMID:30405239 doi:http://dx.doi.org/10.1038/s41586-018-0680-3
- ↑ 3.0 3.1 Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018 Jul;18(7):452-464. doi: 10.1038/s41568-018-0005-8. PMID:29643473 doi:http://dx.doi.org/10.1038/s41568-018-0005-8
- ↑ 4.0 4.1 4.2 4.3 Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B, Buschauer A, Stahlberg H, Altmann KH, Locher KP. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018 Apr;25(4):333-340. doi: 10.1038/s41594-018-0049-1. Epub, 2018 Apr 2. PMID:29610494 doi:http://dx.doi.org/10.1038/s41594-018-0049-1
Student Contributors
Julia Pomeroy